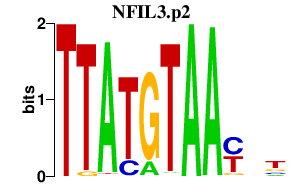
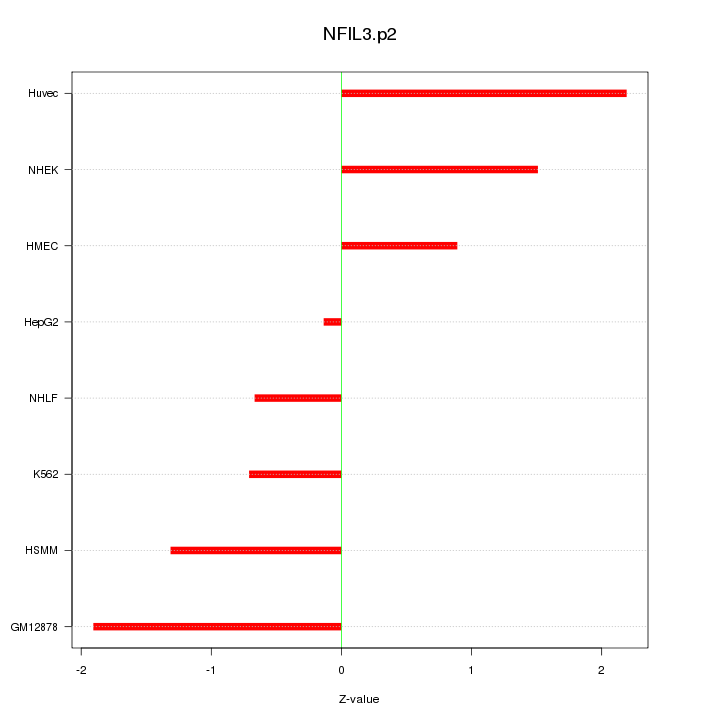
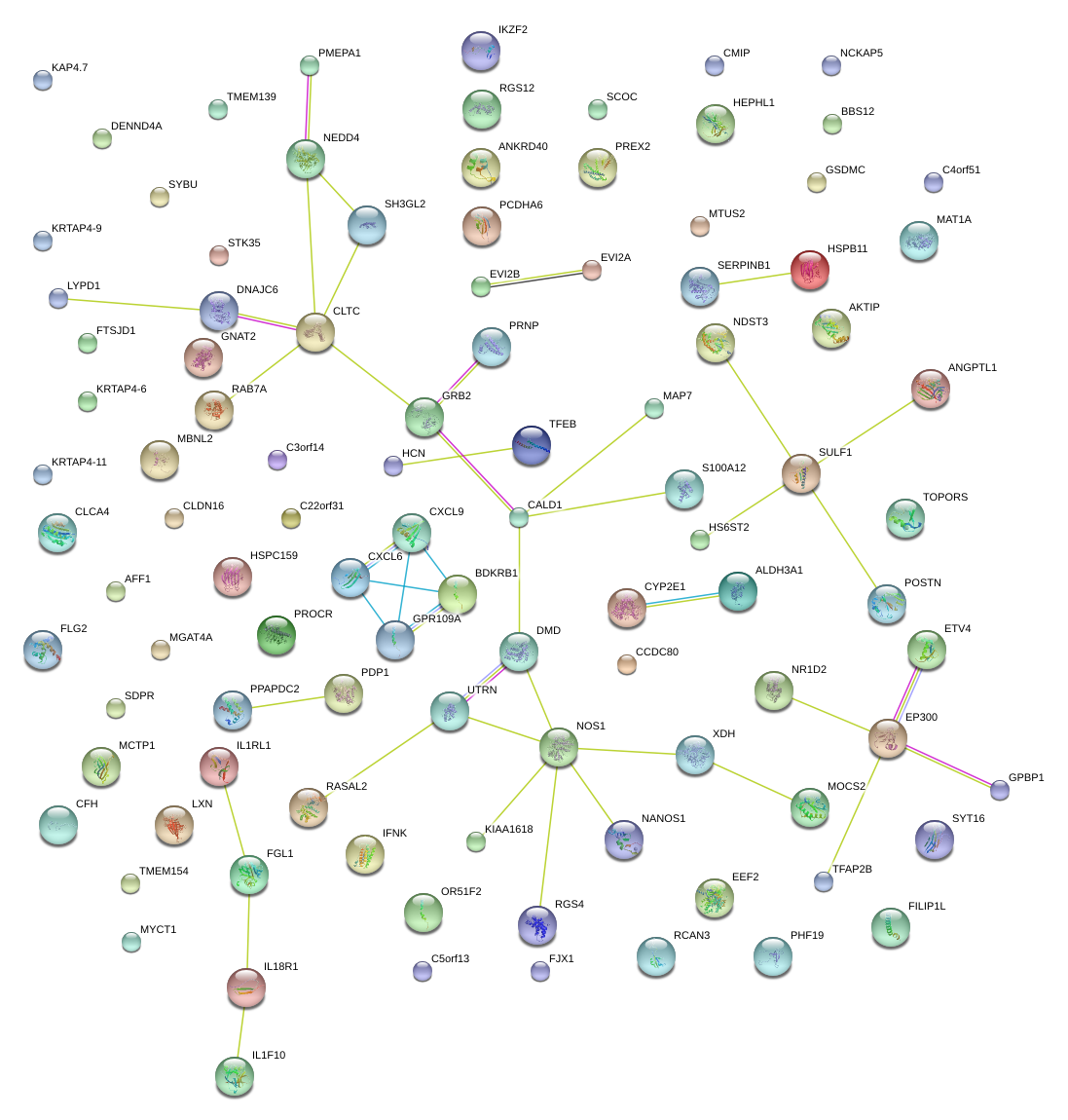
|
chrX_-_33139349
|
1.968
|
NM_004006
|
DMD
|
dystrophin
|
|
chr22_-_34564611
|
1.917
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr6_-_2785682
|
1.912
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr2_-_133895795
|
1.886
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr17_-_19592178
|
1.835
|
NM_000691
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr4_-_77147625
|
1.726
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr8_+_70567634
|
1.687
|
|
SULF1
|
sulfatase 1
|
|
chr2_-_31491102
|
1.656
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr9_+_27514311
|
1.581
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr5_-_111119846
|
1.561
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_31491057
|
1.472
|
|
XDH
|
xanthine dehydrogenase
|
|
chr10_-_82039158
|
1.454
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr2_-_213723298
|
1.426
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr12_-_123568781
|
1.400
|
|
|
|
|
chr4_-_153820535
|
1.376
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr1_+_86785346
|
1.366
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr3_-_159872960
|
1.320
|
NM_020169
|
LXN
|
latexin
|
|
chr19_-_3936460
|
1.294
|
NM_001961
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr4_+_74921136
|
1.288
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr4_+_146820805
|
1.264
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr5_+_56507029
|
1.250
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr14_+_95792299
|
1.242
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr16_-_69876262
|
1.232
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr20_+_2030527
|
1.211
|
NM_080836
|
STK35
|
serine/threonine kinase 35
|
|
chr12_-_116283793
|
1.207
|
NM_000620
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr8_+_70567581
|
1.198
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr2_+_64540843
|
1.195
|
|
HSPC159
|
galectin-related protein
|
|
chr7_+_134114701
|
1.178
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr19_-_3936343
|
1.172
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr3_+_152970904
|
1.167
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr17_+_55098280
|
1.137
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_-_192420168
|
1.100
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr8_+_94999167
|
1.077
|
NM_001161778
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr1_+_194887630
|
1.075
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr12_-_44407515
|
1.056
|
|
LOC400027
|
hypothetical LOC400027
|
|
chr13_+_96726415
|
1.049
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_-_109957195
|
1.047
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr17_-_26665144
|
1.042
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_130014957
|
1.033
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr20_-_55718982
|
1.021
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr17_-_26672757
|
1.020
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr17_+_36515166
|
1.019
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr15_-_63871492
|
0.999
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr9_-_32540818
|
0.985
|
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr5_-_94443300
|
0.971
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr22_+_39817733
|
0.968
|
|
EP300
|
E1A binding protein p300
|
|
chr1_-_151614681
|
0.965
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr21_-_26345294
|
0.913
|
|
|
|
|
chr1_+_65503011
|
0.906
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr17_-_46140015
|
0.890
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr8_-_130868315
|
0.882
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr5_-_94442941
|
0.870
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr17_+_36493927
|
0.864
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr13_-_37070862
|
0.857
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr6_-_136889302
|
0.832
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr6_+_50894397
|
0.830
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr11_+_4799126
|
0.826
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr12_-_44407756
|
0.812
|
|
LOC400027
|
hypothetical LOC400027
|
|
chr4_+_123873306
|
0.797
|
NM_001178007
NM_152618
|
BBS12
|
Bardet-Biedl syndrome 12
|
|
chr20_+_33223434
|
0.790
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr17_-_36528113
|
0.784
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr11_+_65023038
|
0.764
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr2_+_102294393
|
0.764
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr3_-_113842652
|
0.755
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr8_-_17797127
|
0.719
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr3_+_23962518
|
0.703
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_101316038
|
0.696
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr20_+_4614796
|
0.686
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr22_-_27787793
|
0.682
|
NM_015370
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr2_+_113542017
|
0.679
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr9_-_122679387
|
0.665
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr4_+_119174947
|
0.655
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr4_+_88187215
|
0.654
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr8_-_110689396
|
0.649
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_-_52441044
|
0.645
|
NM_004531
|
MOCS2
|
molybdenum cofactor synthesis 2
|
|
chr15_-_53996597
|
0.645
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr13_+_28900776
|
0.633
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr17_-_38979178
|
0.628
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr2_-_133145539
|
0.608
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr1_+_176577228
|
0.604
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr18_-_35634247
|
0.593
|
|
LOC647946
|
hypothetical LOC647946
|
|
chr4_+_3341521
|
0.584
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_-_54183833
|
0.579
|
NM_016126
|
HSPB11
|
heat shock protein family B (small), member 11
|
|
chr11_+_93394025
|
0.574
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr1_-_177106702
|
0.563
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr8_+_69026895
|
0.562
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr3_+_62280435
|
0.561
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr1_+_161305188
|
0.558
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr13_+_96672541
|
0.554
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_+_96672705
|
0.554
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr17_-_70901303
|
0.546
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr6_-_136888771
|
0.544
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr5_+_140235114
|
0.539
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr7_+_142692165
|
0.534
|
NM_153345
|
TMEM139
|
transmembrane protein 139
|
|
chr9_+_17568952
|
0.532
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr16_-_52094613
|
0.532
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr10_+_135190855
|
0.531
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr2_-_98646367
|
0.531
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_-_150599105
|
0.527
|
NM_001014342
|
FLG2
|
filaggrin family member 2
|
|
chr1_+_24713396
|
0.520
|
|
RCAN3
|
RCAN family member 3
|
|
chr3_+_191588354
|
0.514
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr4_+_141397889
|
0.513
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr6_-_41810712
|
0.512
|
NM_007162
|
TFEB
|
transcription factor EB
|
|
chr11_+_35596310
|
0.511
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chrX_+_56772391
|
0.484
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr16_+_80086454
|
0.484
|
NM_030629
|
CMIP
|
c-Maf-inducing protein
|
|
chr9_+_674193
|
0.480
|
|
|
|
|
chr14_+_61532293
|
0.474
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chrX_-_131919896
|
0.469
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr9_-_122679283
|
0.468
|
|
PHF19
|
PHD finger protein 19
|
|
chr12_-_121753795
|
0.459
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr6_+_153060722
|
0.443
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr4_-_74307637
|
0.435
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr5_-_139993469
|
0.434
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr2_+_87536106
|
0.427
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr14_+_101345878
|
0.423
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr1_+_149165438
|
0.417
|
NM_001145415
NM_012432
|
SETDB1
|
SET domain, bifurcated 1
|
|
chr9_+_15542871
|
0.414
|
NM_173550
|
C9orf93
|
chromosome 9 open reading frame 93
|
|
chr18_+_30875297
|
0.411
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr1_+_195213289
|
0.409
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr17_-_64649608
|
0.408
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr12_+_50633430
|
0.406
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr17_+_7402788
|
0.405
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr2_-_104834344
|
0.398
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr10_-_119795663
|
0.392
|
NM_014904
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chr1_+_151081953
|
0.390
|
NM_001128600
|
LCE6A
|
late cornified envelope 6A
|
|
chr6_-_64087806
|
0.385
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr17_-_64752550
|
0.383
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr5_-_56814310
|
0.380
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr7_+_80145461
|
0.370
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_-_245763763
|
0.369
|
NM_198074
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3
|
|
chr17_-_36074812
|
0.365
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr3_+_103029523
|
0.363
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr6_-_134415311
|
0.357
|
NM_145176
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12
|
|
chr17_-_31337798
|
0.354
|
NM_032962
NM_032963
|
CCL14
|
chemokine (C-C motif) ligand 14
|
|
chr4_-_70760381
|
0.354
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr8_-_102286695
|
0.353
|
|
ZNF706
|
zinc finger protein 706
|
|
chr4_-_87989291
|
0.349
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr12_-_7487989
|
0.346
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr17_-_59204622
|
0.344
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr4_+_110574420
|
0.342
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr17_-_36559579
|
0.341
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr19_+_56671649
|
0.340
|
NM_001080405
|
CEACAM18
|
carcinoembryonic antigen-related cell adhesion molecule 18
|
|
chr2_+_87536174
|
0.340
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr10_+_4995453
|
0.337
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr8_-_103735194
|
0.336
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr9_-_98104206
|
0.334
|
NM_000197
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr3_-_192063158
|
0.329
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr11_-_128399218
|
0.329
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_+_57093030
|
0.324
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr17_-_26665194
|
0.321
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr2_-_45648608
|
0.319
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr12_+_110328126
|
0.318
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_+_144236346
|
0.317
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr11_+_111552298
|
0.314
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr8_-_11891168
|
0.313
|
NM_001033019
|
DEFB134
|
defensin, beta 134
|
|
chr5_+_36642424
|
0.311
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr20_+_36643251
|
0.309
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chr4_+_81325447
|
0.307
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr5_-_158567411
|
0.306
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr8_+_22075112
|
0.298
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr5_+_36642213
|
0.291
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_-_73889777
|
0.284
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr10_-_102663070
|
0.283
|
|
|
|
|
chr8_-_102287083
|
0.282
|
NM_001042510
NM_016096
|
ZNF706
|
zinc finger protein 706
|
|
chr5_-_79982584
|
0.282
|
NM_001190470
|
MTRNR2L2
|
MT-RNR2-like 2
|
|
chr2_-_188127294
|
0.280
|
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr21_+_18539016
|
0.279
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr7_+_89681069
|
0.276
|
NM_001040666
|
STEAP2
|
six transmembrane epithelial antigen of the prostate 2
|
|
chr7_+_20653490
|
0.275
|
NM_001163942
NM_001163993
NM_178559
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr2_-_188021117
|
0.274
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr5_-_139992621
|
0.273
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr6_-_50045296
|
0.272
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chrX_-_84521052
|
0.266
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr5_-_38592439
|
0.265
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr4_-_24590867
|
0.264
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr15_+_56217686
|
0.263
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr10_+_4995597
|
0.261
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr4_-_96689373
|
0.261
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chrX_+_38548316
|
0.258
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr1_-_42157077
|
0.255
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr7_+_150321771
|
0.253
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr1_+_144236398
|
0.253
|
|
ITGA10
|
integrin, alpha 10
|
|
chr2_+_135392857
|
0.253
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr8_-_92122219
|
0.252
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr17_-_26665220
|
0.251
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_44667504
|
0.251
|
|
ZNF35
|
zinc finger protein 35
|
|
chr4_+_120167264
|
0.250
|
|
SYNPO2
|
synaptopodin 2
|
|
chr10_+_91077581
|
0.250
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr2_+_135392879
|
0.247
|
|
CCNT2
|
cyclin T2
|
|
chr12_-_69317441
|
0.247
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr1_-_114231519
|
0.247
|
|
BCL2L15
|
BCL2-like 15
|
|
chr5_-_14924555
|
0.246
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr3_-_102194938
|
0.244
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chrX_+_117992603
|
0.244
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_+_38548016
|
0.243
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr10_-_99780336
|
0.242
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr7_-_31347032
|
0.239
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr14_-_69333624
|
0.237
|
NM_003049
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr12_-_28016182
|
0.234
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|