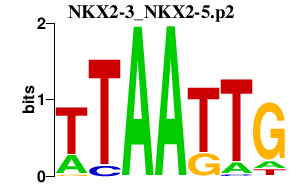
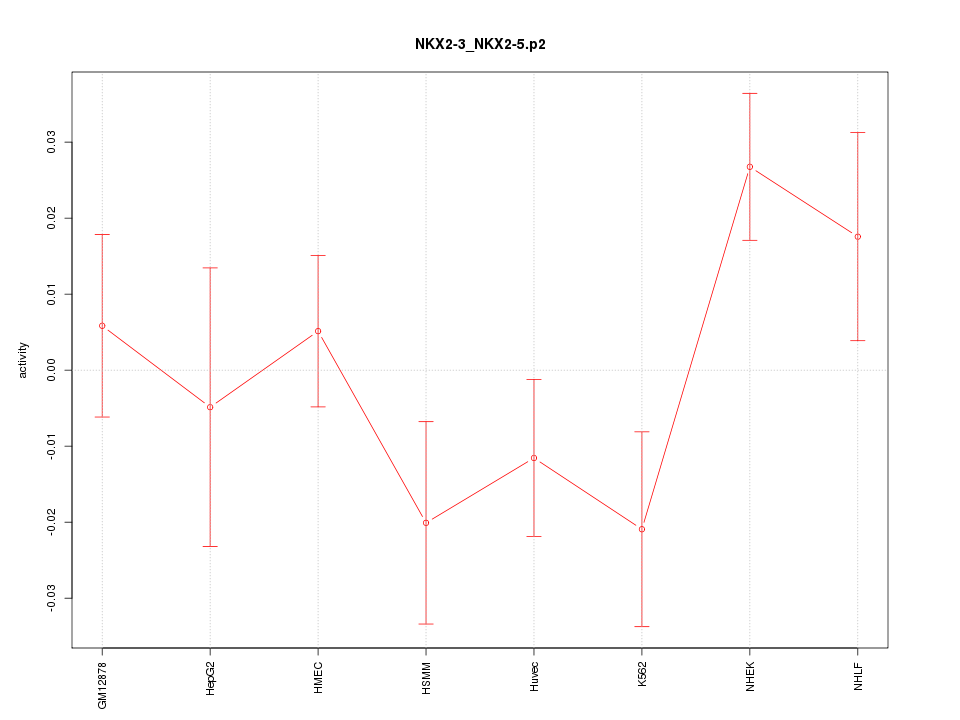
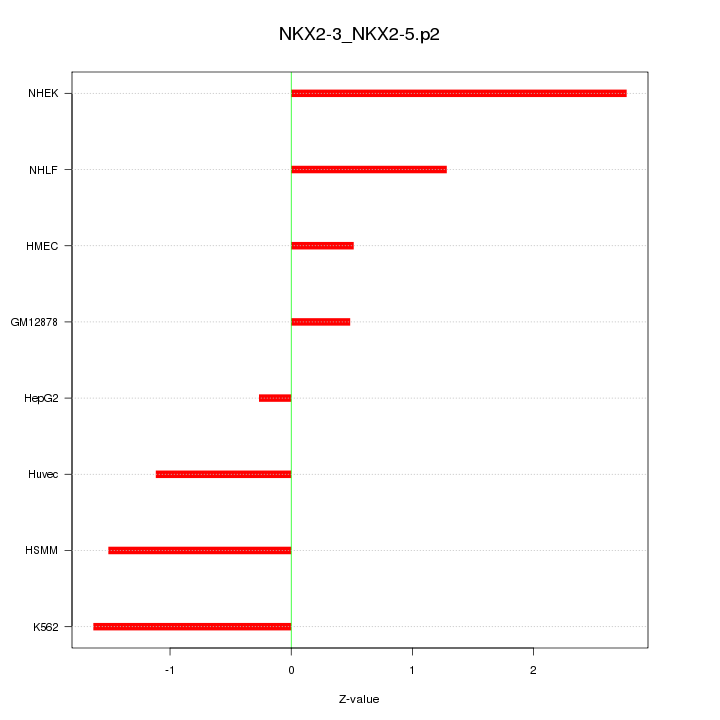
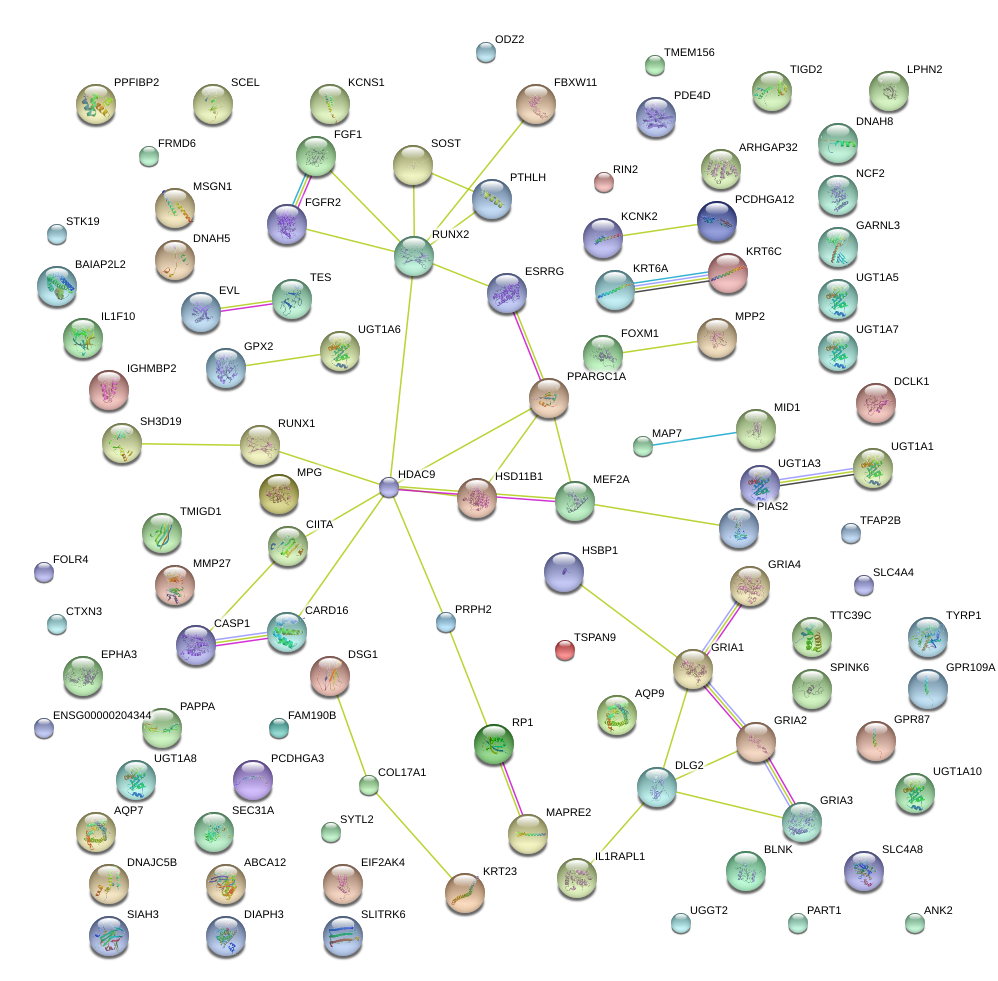
|
chr5_-_13997588
|
2.619
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr10_-_105835609
|
1.967
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr10_-_105835525
|
1.957
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr15_-_64108718
|
1.942
|
|
|
|
|
chr3_+_89239494
|
1.875
|
|
EPHA3
|
EPH receptor A3
|
|
chr11_+_7574992
|
1.841
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr13_-_45323757
|
1.839
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr18_+_27152049
|
1.832
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr3_-_152517204
|
1.757
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr14_+_99633570
|
1.730
|
|
EVL
|
Enah/Vasp-like
|
|
chr10_-_116241572
|
1.678
|
|
|
|
|
chr15_+_97990705
|
1.671
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr4_-_152368492
|
1.663
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_+_127012611
|
1.644
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr18_+_19826734
|
1.606
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr4_+_114433551
|
1.543
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr1_+_207944807
|
1.530
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr11_-_104411065
|
1.530
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr1_-_181826633
|
1.523
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr10_-_98021144
|
1.519
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_37282516
|
1.465
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chr16_+_10878539
|
1.393
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr17_-_25685175
|
1.391
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr13_-_85271329
|
1.346
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr20_-_43163021
|
1.275
|
NM_002251
|
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1
|
|
chr13_-_59485299
|
1.269
|
NM_030932
|
DIAPH3
|
diaphanous homolog 3 (Drosophila)
|
|
chr1_-_214963279
|
1.265
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr13_-_35327798
|
1.259
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr13_+_77007809
|
1.239
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr17_-_36347197
|
1.192
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_+_17861266
|
1.187
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr8_+_67096344
|
1.152
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr14_+_51025604
|
1.150
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr5_+_59819761
|
1.117
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr11_-_128567302
|
1.114
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr21_-_35343065
|
1.063
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr5_+_140848905
|
1.055
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr11_-_104477367
|
1.047
|
NM_001007232
|
CARD17
|
caspase recruitment domain family, member 17
|
|
chr1_+_213245507
|
1.027
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr9_+_129066576
|
1.021
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr1_+_81544432
|
1.020
|
|
LPHN2
|
latrophilin 2
|
|
chr10_-_123346148
|
1.008
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr9_+_117955890
|
0.988
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr7_+_18502342
|
0.981
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr5_+_166644420
|
0.972
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr12_-_51173238
|
0.949
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr7_+_115650240
|
0.942
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr11_+_93678450
|
0.940
|
NM_001080486
|
FOLR4
|
folate receptor 4 (delta) homolog (mouse)
|
|
chr17_-_39191658
|
0.932
|
NM_025237
|
SOST
|
sclerostin
|
|
chr11_-_85115105
|
0.930
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr10_+_86174665
|
0.926
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr4_-_23500706
|
0.908
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr5_-_141973875
|
0.907
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_36346240
|
0.906
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_-_102081654
|
0.890
|
NM_022122
|
MMP27
|
matrix metallopeptidase 27
|
|
chr12_-_121753795
|
0.882
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr15_+_38082041
|
0.874
|
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr2_-_215711313
|
0.868
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr12_-_121767272
|
0.860
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr8_+_55691179
|
0.856
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chrX_+_28515479
|
0.854
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr9_+_12683385
|
0.847
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr6_+_50894397
|
0.847
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr12_-_28016182
|
0.842
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr5_+_152850276
|
0.837
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr5_+_147562531
|
0.832
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr4_+_90252990
|
0.821
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr14_-_64479198
|
0.821
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr5_-_59819642
|
0.821
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_-_36836620
|
0.820
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr4_+_72423633
|
0.813
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_38710376
|
0.808
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr2_+_234266250
|
0.807
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr11_-_83071084
|
0.806
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr22_-_36836478
|
0.804
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr2_+_113546365
|
0.797
|
NM_032556
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chrX_-_10504852
|
0.793
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr3_+_89239363
|
0.785
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr20_+_19863716
|
0.776
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_-_136888771
|
0.774
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr15_+_56217686
|
0.767
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr10_-_31328426
|
0.761
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr1_+_156229686
|
0.753
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr1_+_208568218
|
0.740
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr6_+_134800533
|
0.731
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr9_-_116305273
|
0.730
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr8_-_122722674
|
0.728
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr12_-_16650687
|
0.721
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_161981138
|
0.718
|
|
TBR1
|
T-box, brain, 1
|
|
chr14_+_22059212
|
0.718
|
|
|
|
|
chr10_+_101532544
|
0.713
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr15_-_59308708
|
0.707
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_+_101532450
|
0.705
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr1_+_182040837
|
0.700
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr4_+_3341521
|
0.700
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chrX_-_64112993
|
0.699
|
NM_001178033
NM_018684
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr5_-_22889487
|
0.699
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr12_-_16650697
|
0.695
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_139315011
|
0.694
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr4_-_87593306
|
0.692
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chrX_+_21960841
|
0.686
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr15_-_68781662
|
0.684
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_+_13903936
|
0.683
|
NM_012231
NM_015866
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr1_-_181889070
|
0.674
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr17_+_72982919
|
0.672
|
NM_001113495
|
SEPT9
|
septin 9
|
|
chr13_-_46368968
|
0.671
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr7_+_113842284
|
0.671
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr1_+_154520693
|
0.657
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr14_+_22059174
|
0.655
|
|
|
|
|
chr18_+_30544193
|
0.655
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr13_+_96726415
|
0.653
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr2_+_161981046
|
0.652
|
|
TBR1
|
T-box, brain, 1
|
|
chr13_-_37070862
|
0.650
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr11_-_123353607
|
0.644
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr6_+_136214495
|
0.641
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr15_+_75074793
|
0.636
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chrX_+_66705407
|
0.634
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr4_-_68432293
|
0.631
|
NM_004262
|
TMPRSS11D
|
transmembrane protease, serine 11D
|
|
chr10_+_95507555
|
0.627
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr4_+_68995761
|
0.625
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr1_+_221168388
|
0.625
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr3_-_101316038
|
0.623
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr4_-_21308415
|
0.622
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr20_-_49747397
|
0.621
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr2_+_113542017
|
0.620
|
NM_173161
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr4_-_83566045
|
0.619
|
|
|
|
|
chr12_+_79362256
|
0.617
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr10_+_102113410
|
0.614
|
|
|
|
|
chr7_+_18501876
|
0.614
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr6_+_71192862
|
0.612
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr12_-_14024187
|
0.611
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr12_+_9871343
|
0.611
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr2_+_161980865
|
0.609
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr15_-_59308770
|
0.605
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_+_75074743
|
0.601
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr10_+_80743769
|
0.597
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr3_-_11598829
|
0.596
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr16_+_20682812
|
0.595
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr1_+_185064654
|
0.594
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr6_-_52882419
|
0.594
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr3_-_110155237
|
0.592
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr15_-_53996597
|
0.592
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr16_+_22432344
|
0.585
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr11_-_117305277
|
0.581
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr15_+_38849450
|
0.579
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr1_-_57057956
|
0.575
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chr21_-_42219849
|
0.561
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr10_+_90552466
|
0.561
|
NM_001128215
|
LIPM
|
lipase, family member M
|
|
chr3_+_63928459
|
0.551
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chrX_+_105823723
|
0.549
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr9_-_33455191
|
0.545
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr12_-_10173947
|
0.544
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr5_-_39255335
|
0.543
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr10_-_105427898
|
0.540
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_57242492
|
0.539
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr10_+_24795446
|
0.534
|
|
KIAA1217
|
KIAA1217
|
|
chr12_+_39372456
|
0.532
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr12_+_8866416
|
0.532
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr20_-_33489383
|
0.531
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chrX_-_133620103
|
0.524
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr17_+_56843893
|
0.524
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr17_-_19290745
|
0.523
|
|
|
|
|
chr3_+_2528272
|
0.521
|
|
CNTN4
|
contactin 4
|
|
chr19_+_42689881
|
0.519
|
|
ZNF793
|
zinc finger protein 793
|
|
chr3_+_101316463
|
0.519
|
NM_001167924
|
C3orf26
|
chromosome 3 open reading frame 26
|
|
chr7_-_112513379
|
0.518
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr4_-_83566246
|
0.517
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr7_-_27133163
|
0.516
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr20_-_62052922
|
0.515
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr3_+_160269734
|
0.514
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr1_-_155336223
|
0.512
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr11_+_117452995
|
0.510
|
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr4_-_116254381
|
0.506
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr21_-_38955487
|
0.500
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr13_-_19703371
|
0.500
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr18_-_26996742
|
0.500
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr20_+_9236446
|
0.499
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr2_-_40593074
|
0.496
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chrX_+_111005934
|
0.496
|
NM_001195576
NM_001195578
|
LOC100329135
|
hypothetical LOC100329135
|
|
chr7_+_80113903
|
0.494
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr4_-_176970895
|
0.493
|
|
GPM6A
|
glycoprotein M6A
|
|
chr21_-_35343331
|
0.491
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_-_176971175
|
0.490
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr14_+_51397134
|
0.488
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr8_-_110689396
|
0.488
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_+_126378507
|
0.486
|
|
NCRNA00288
|
non-protein coding RNA 288
|
|
chrX_-_43717864
|
0.486
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr1_-_42156913
|
0.484
|
|
|
|
|
chr8_-_102872614
|
0.477
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr12_-_6354650
|
0.476
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr3_+_42165740
|
0.475
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr3_+_42165829
|
0.471
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr5_+_140772691
|
0.470
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr8_-_67253234
|
0.468
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr7_-_87694218
|
0.466
|
NM_198901
|
SRI
|
sorcin
|
|
chr17_+_71232370
|
0.463
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr12_+_28301399
|
0.462
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr18_+_18989786
|
0.462
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr12_+_54400417
|
0.462
|
NM_002905
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis)
|
|
chr1_+_50342168
|
0.451
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|