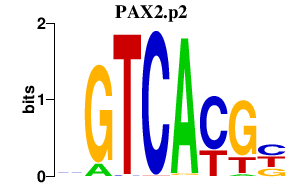
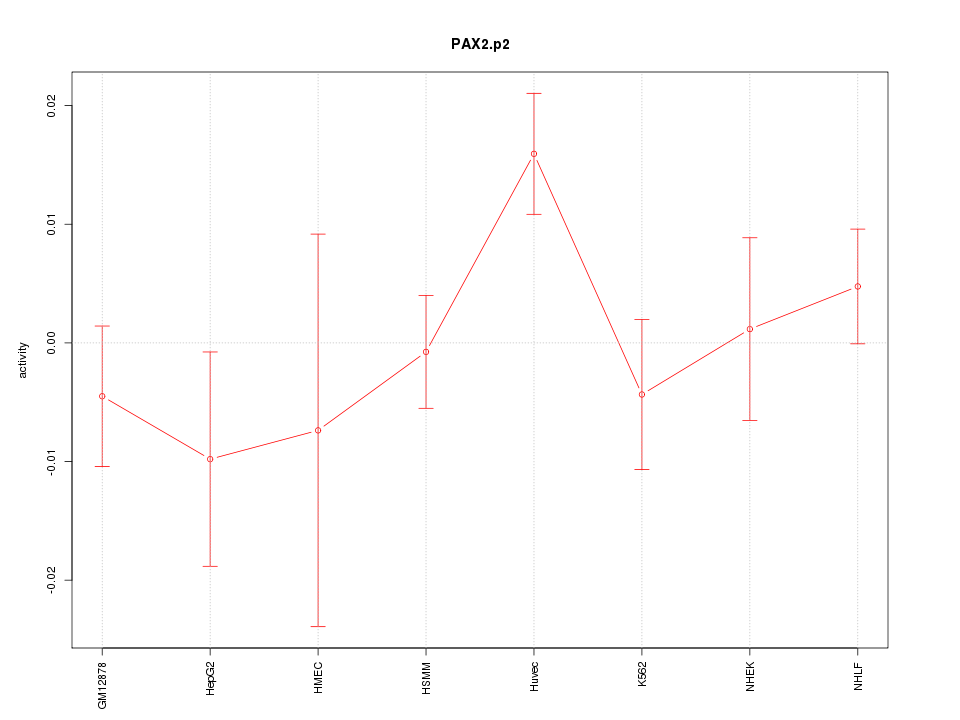
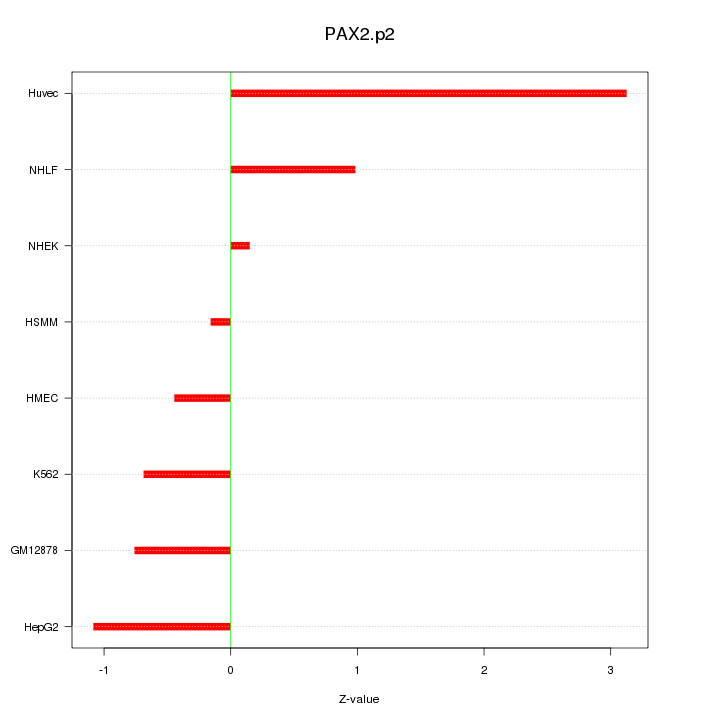
|
chr15_-_68781662
|
1.269
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_-_154205675
|
1.228
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr17_-_32730804
|
0.976
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr12_+_50592379
|
0.963
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr1_-_11788666
|
0.905
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr5_-_16795336
|
0.857
|
|
MYO10
|
myosin X
|
|
chr22_+_39583001
|
0.851
|
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chrX_+_55760774
|
0.822
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr1_-_197173159
|
0.813
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr1_-_171286634
|
0.781
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chrX_+_101862297
|
0.762
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr10_-_14090459
|
0.750
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr6_-_84994032
|
0.748
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr8_-_82557956
|
0.736
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr15_-_63871492
|
0.719
|
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr17_+_7416780
|
0.706
|
NM_001416
|
EIF4A1
|
eukaryotic translation initiation factor 4A1
|
|
chr6_-_43135092
|
0.696
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr1_-_109957195
|
0.692
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr3_-_99724047
|
0.678
|
|
CLDND1
|
claudin domain containing 1
|
|
chr1_-_11788550
|
0.670
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr5_-_167938875
|
0.669
|
|
PANK3
|
pantothenate kinase 3
|
|
chr5_+_14493905
|
0.665
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr12_-_121316907
|
0.659
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr3_+_67131416
|
0.656
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr8_-_13416554
|
0.654
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr17_+_70984173
|
0.647
|
|
KIAA0195
|
KIAA0195
|
|
chr15_-_71712711
|
0.614
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr9_-_126573349
|
0.602
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr1_-_202595666
|
0.598
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr4_+_78298326
|
0.597
|
|
CCNG2
|
cyclin G2
|
|
chr5_-_167938991
|
0.595
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr16_+_56219802
|
0.591
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr4_+_81337663
|
0.591
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr14_-_64040072
|
0.590
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr7_+_150442652
|
0.585
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr7_-_27162688
|
0.581
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr5_+_148186348
|
0.574
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr2_+_219751230
|
0.573
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr5_-_148422799
|
0.572
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr11_-_85457469
|
0.572
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_8993460
|
0.570
|
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr11_+_125586828
|
0.560
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr21_+_29439768
|
0.533
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_+_105320440
|
0.533
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr1_+_221168388
|
0.531
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr18_+_9126742
|
0.530
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr5_+_172415891
|
0.529
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr14_+_76634323
|
0.524
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr15_-_63871598
|
0.515
|
NM_001144823
NM_005848
|
DENND4A
|
DENN/MADD domain containing 4A
|
|
chr14_-_67353046
|
0.514
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr12_-_122415708
|
0.507
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr11_-_18300246
|
0.504
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr5_-_159730196
|
0.503
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chr3_+_190147633
|
0.502
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr3_+_11153778
|
0.499
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr9_-_90983501
|
0.498
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr6_+_43135346
|
0.494
|
NM_138343
NM_201521
|
KLC4
|
kinesin light chain 4
|
|
chr5_+_41940216
|
0.490
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr1_+_11788889
|
0.490
|
|
CLCN6
|
chloride channel 6
|
|
chr12_-_8993515
|
0.487
|
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr5_-_71838875
|
0.486
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr15_-_71712605
|
0.485
|
|
NPTN
|
neuroplastin
|
|
chr20_+_37024383
|
0.483
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr7_-_81237162
|
0.482
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr5_-_141372721
|
0.478
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr17_+_75766875
|
0.478
|
NM_024110
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr5_-_90645974
|
0.478
|
|
LOC100505994
|
hypothetical LOC100505994
|
|
chr14_-_19999324
|
0.474
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr17_-_43977273
|
0.464
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr9_+_70978790
|
0.463
|
NM_001170630
NM_004817
NM_201629
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr4_+_144325533
|
0.462
|
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr9_-_37566248
|
0.461
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr1_-_77921681
|
0.460
|
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr19_+_12709281
|
0.460
|
NM_004317
|
ASNA1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial)
|
|
chr1_-_77920914
|
0.456
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr1_-_152797707
|
0.455
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr4_+_120029426
|
0.452
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr12_-_56432292
|
0.451
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_+_11788793
|
0.447
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr17_-_38081967
|
0.444
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr12_-_121316997
|
0.444
|
NM_022916
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr12_-_56432377
|
0.443
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr10_+_22645370
|
0.437
|
|
COMMD3
|
COMM domain containing 3
|
|
chr20_-_17487480
|
0.436
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr3_+_114948550
|
0.435
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr7_-_105712646
|
0.435
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr15_-_73719546
|
0.433
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr19_-_17236522
|
0.433
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr1_+_32485404
|
0.431
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr4_+_144325502
|
0.431
|
NM_032557
|
USP38
|
ubiquitin specific peptidase 38
|
|
chr11_-_125586733
|
0.421
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr14_-_34943574
|
0.420
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr6_+_87921980
|
0.418
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr6_+_148705421
|
0.417
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr12_-_101115677
|
0.414
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr10_-_31360778
|
0.414
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr10_+_45542653
|
0.412
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr8_-_102034370
|
0.406
|
NM_001135699
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr6_+_127629677
|
0.405
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr12_+_15926681
|
0.403
|
|
STRAP
|
serine/threonine kinase receptor associated protein
|
|
chr9_+_102380156
|
0.402
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr14_+_73621408
|
0.402
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr8_-_102034777
|
0.397
|
NM_003406
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr22_-_37425937
|
0.396
|
NM_014876
|
JOSD1
|
Josephin domain containing 1
|
|
chr17_+_4743645
|
0.394
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr1_+_113300507
|
0.391
|
|
LOC100506392
|
hypothetical LOC100506392
|
|
chr11_+_46325457
|
0.389
|
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr11_-_113251415
|
0.388
|
NM_020886
|
USP28
|
ubiquitin specific peptidase 28
|
|
chr9_-_90983195
|
0.388
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr9_+_119506446
|
0.386
|
|
TLR4
|
toll-like receptor 4
|
|
chr12_+_54396118
|
0.386
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr7_+_102340579
|
0.386
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr18_-_44729700
|
0.385
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr11_-_85457736
|
0.385
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr17_+_6856676
|
0.384
|
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chr11_+_18300689
|
0.384
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr10_+_70553862
|
0.382
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr20_+_19863716
|
0.381
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr17_+_6856697
|
0.381
|
|
RNASEK-C17ORF49
|
RNASEK-C17orf49 read-through transcript
|
|
chr10_+_70609986
|
0.380
|
NM_003171
|
SUPV3L1
|
suppressor of var1, 3-like 1 (S. cerevisiae)
|
|
chr4_-_38710376
|
0.379
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr10_+_22645304
|
0.379
|
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 read-through transcript
COMM domain containing 3
|
|
chr1_+_11788827
|
0.378
|
|
CLCN6
|
chloride channel 6
|
|
chr17_-_46692369
|
0.377
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr6_+_143423656
|
0.377
|
NM_016108
|
AIG1
|
androgen-induced 1
|
|
chr9_-_21999270
|
0.376
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr10_+_128583984
|
0.376
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr1_-_184916044
|
0.375
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr10_+_101532450
|
0.371
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr17_+_3993690
|
0.371
|
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr12_-_56432385
|
0.370
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr4_-_90197140
|
0.370
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr6_-_109810396
|
0.366
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr19_-_50912429
|
0.364
|
|
|
|
|
chr2_+_219751207
|
0.360
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr2_-_225519974
|
0.359
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr4_+_93444572
|
0.358
|
NM_001510
|
GRID2
|
glutamate receptor, ionotropic, delta 2
|
|
chr3_+_114948600
|
0.358
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr12_+_12829807
|
0.358
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr6_-_37061924
|
0.358
|
NM_014341
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr7_-_103635587
|
0.356
|
NM_181747
NM_002553
|
ORC5
|
origin recognition complex, subunit 5
|
|
chr10_-_3817418
|
0.356
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr10_+_102662675
|
0.356
|
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr16_+_1341937
|
0.356
|
|
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit
|
|
chr3_-_198509276
|
0.355
|
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr8_-_102034272
|
0.353
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr5_+_34720368
|
0.352
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr9_+_35782405
|
0.351
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr9_+_35528628
|
0.351
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr19_-_45914618
|
0.351
|
NM_024876
|
ADCK4
|
aarF domain containing kinase 4
|
|
chr17_+_6879960
|
0.351
|
NM_201566
|
SLC16A13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13)
|
|
chr12_-_56432375
|
0.351
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr10_-_112054497
|
0.351
|
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr22_+_40195058
|
0.351
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr18_+_53863865
|
0.351
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_+_34066679
|
0.347
|
|
ALG10
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe)
|
|
chr14_+_54807855
|
0.344
|
|
FBXO34
|
F-box protein 34
|
|
chr22_-_30671298
|
0.344
|
NM_015372
|
C22orf24
|
chromosome 22 open reading frame 24
|
|
chr11_-_85115105
|
0.342
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr1_+_15725894
|
0.341
|
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr10_-_45410197
|
0.341
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr2_+_108571209
|
0.339
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr18_+_52469559
|
0.339
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr4_+_113372287
|
0.339
|
NM_001128426
NM_018569
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein
|
|
chr1_+_36462625
|
0.339
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr6_-_112515329
|
0.338
|
NM_016262
|
TUBE1
|
tubulin, epsilon 1
|
|
chr4_-_177950658
|
0.337
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chrX_+_24077649
|
0.337
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr2_+_108571177
|
0.336
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_+_98725505
|
0.336
|
NM_178812
|
MTDH
|
metadherin
|
|
chr5_-_94916416
|
0.333
|
NM_014639
|
TTC37
|
tetratricopeptide repeat domain 37
|
|
chr6_+_17501589
|
0.333
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr14_-_49652944
|
0.332
|
NM_001040662
NM_024558
|
METTL21D
|
methyltransferase like 21D
|
|
chr21_-_35343456
|
0.332
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_+_107598960
|
0.330
|
|
ATM
|
ataxia telangiectasia mutated
|
|
chr8_-_130868315
|
0.329
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr2_+_219751192
|
0.327
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr14_-_76612286
|
0.327
|
|
|
|
|
chr5_-_75954807
|
0.326
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr3_-_20202622
|
0.325
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr9_+_70978970
|
0.324
|
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr15_+_43667050
|
0.323
|
|
PLDN
|
pallidin homolog (mouse)
|
|
chr11_+_66915997
|
0.323
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr2_-_216944911
|
0.321
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr7_+_92699588
|
0.321
|
NM_017667
NM_024553
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr12_+_88627601
|
0.321
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr3_+_101462375
|
0.321
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr2_+_108571336
|
0.321
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr10_-_112054679
|
0.320
|
NM_005871
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr8_+_120148604
|
0.319
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr3_+_52419713
|
0.317
|
|
PHF7
|
PHD finger protein 7
|
|
chr3_-_123995216
|
0.317
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr10_+_22649977
|
0.316
|
NM_005180
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr1_+_15725935
|
0.316
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr9_-_37894098
|
0.316
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr1_+_168030798
|
0.316
|
|
C1orf112
|
chromosome 1 open reading frame 112
|
|
chr3_+_23962518
|
0.314
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr6_-_151754337
|
0.314
|
NM_020861
|
ZBTB2
|
zinc finger and BTB domain containing 2
|
|
chr3_-_198509808
|
0.313
|
NM_001098424
NM_004087
|
DLG1
|
discs, large homolog 1 (Drosophila)
|
|
chr1_+_114273518
|
0.312
|
NM_152696
NM_198268
|
HIPK1
|
homeodomain interacting protein kinase 1
|