|
chr11_-_16380967
|
5.576
|
NM_001145819
NM_017508
|
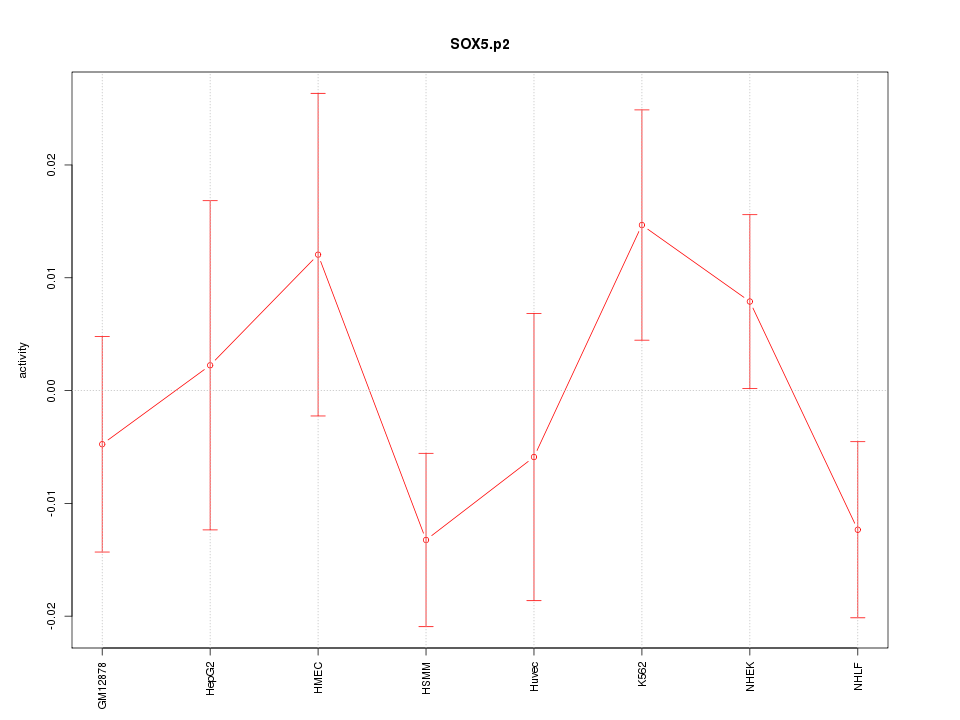
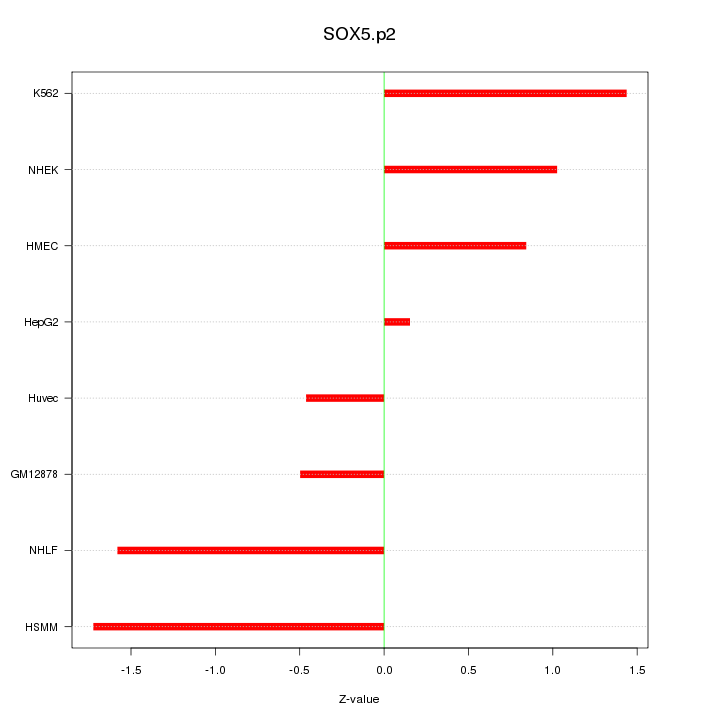
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr7_+_128802719
|
2.521
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_123353607
|
2.408
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr8_-_38324809
|
2.099
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr7_-_16471817
|
2.065
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr21_+_42492867
|
1.703
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr1_+_207668787
|
1.423
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr11_+_20005704
|
1.420
|
|
NAV2
|
neuron navigator 2
|
|
chr6_-_29752909
|
1.294
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr17_+_63252704
|
1.283
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr19_-_56164628
|
1.281
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr6_-_49712483
|
1.242
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr1_+_116727514
|
1.236
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr10_-_32707549
|
1.230
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr2_+_169631777
|
1.229
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_-_191422346
|
1.204
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr1_-_198643885
|
1.197
|
|
ZNF281
|
zinc finger protein 281
|
|
chr17_-_71285992
|
1.197
|
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr2_-_160181206
|
1.163
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr10_+_5976354
|
1.126
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr13_-_83354425
|
1.076
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr9_+_126460511
|
1.073
|
|
|
|
|
chr9_+_134884627
|
1.069
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr2_-_157890400
|
1.063
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr12_+_68028397
|
1.056
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr4_-_76817635
|
1.050
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr5_-_147266257
|
1.045
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr1_-_31117948
|
1.037
|
|
SDC3
|
syndecan 3
|
|
chr3_+_46591048
|
1.036
|
NM_001174136
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr12_+_68028407
|
1.028
|
|
LYZ
|
lysozyme
|
|
chr17_-_36996611
|
1.027
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr13_-_83354474
|
1.003
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr6_+_29902734
|
1.002
|
NM_002127
|
HLA-G
|
major histocompatibility complex, class I, G
|
|
chrX_-_151370325
|
0.978
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr6_+_12398514
|
0.931
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr17_+_63252668
|
0.921
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr1_+_151270301
|
0.913
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr3_+_138159396
|
0.912
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr1_-_150564302
|
0.896
|
NM_002016
|
FLG
|
filaggrin
|
|
chr3_-_57209319
|
0.868
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr7_-_26198673
|
0.865
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr20_-_60435983
|
0.851
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr5_-_148738980
|
0.843
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr4_-_40211324
|
0.842
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr11_+_56224436
|
0.838
|
NM_001005213
NM_001013358
|
OR9G1
OR9G9
|
olfactory receptor, family 9, subfamily G, member 1
olfactory receptor, family 9, subfamily G, member 9
|
|
chr1_-_198644111
|
0.828
|
|
ZNF281
|
zinc finger protein 281
|
|
chr1_-_26070330
|
0.819
|
NM_178422
|
PAQR7
|
progestin and adipoQ receptor family member VII
|
|
chr10_+_63331018
|
0.811
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chrX_+_70419766
|
0.806
|
NM_001145408
NM_001145409
NM_001145410
NM_007363
|
NONO
|
non-POU domain containing, octamer-binding
|
|
chr1_-_108032645
|
0.784
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_-_166149867
|
0.775
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr17_-_53847937
|
0.773
|
|
RNF43
|
ring finger protein 43
|
|
chr9_-_85773899
|
0.771
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr9_-_129752687
|
0.770
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr7_-_16811131
|
0.761
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr1_+_148603208
|
0.734
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr10_+_63331401
|
0.730
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_-_21375891
|
0.704
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr1_-_16411690
|
0.702
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr1_+_166516901
|
0.701
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr1_+_183281173
|
0.700
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chr20_+_31613800
|
0.696
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr3_-_150993299
|
0.690
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr1_+_165565006
|
0.689
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_+_156128431
|
0.685
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chrX_-_116991728
|
0.683
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chrX_+_135441970
|
0.680
|
NM_016267
|
VGLL1
|
vestigial like 1 (Drosophila)
|
|
chr5_-_121842680
|
0.662
|
|
|
|
|
chr6_-_116973465
|
0.660
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr1_-_21375939
|
0.658
|
NM_001198801
NM_001198802
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr10_-_105835609
|
0.655
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr11_-_4555625
|
0.648
|
NM_144663
|
C11orf40
|
chromosome 11 open reading frame 40
|
|
chr18_+_59405513
|
0.640
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr2_-_99238001
|
0.637
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr6_-_135312899
|
0.636
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr1_+_165564904
|
0.633
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr22_+_38652555
|
0.630
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr2_+_62787534
|
0.618
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr19_+_12810258
|
0.614
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr2_-_225073357
|
0.612
|
|
CUL3
|
cullin 3
|
|
chr6_-_41276901
|
0.600
|
NM_024807
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chrX_+_105823723
|
0.596
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr20_+_58004811
|
0.583
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chrX_+_15435351
|
0.578
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr8_-_11742668
|
0.575
|
|
|
|
|
chr12_-_23995220
|
0.574
|
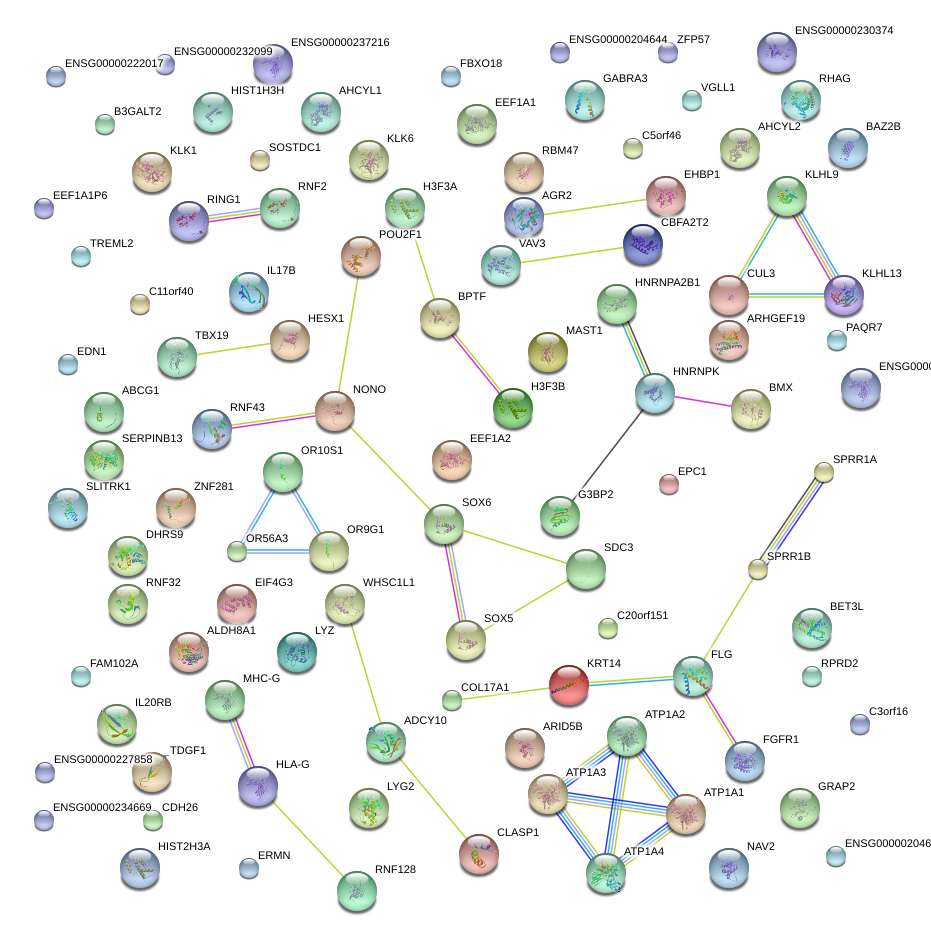
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_+_5925152
|
0.565
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr11_+_65026382
|
0.555
|
|
|
|
|
chr10_+_49279647
|
0.550
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr11_+_64950074
|
0.540
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr3_+_182900081
|
0.538
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr10_+_49184658
|
0.538
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr17_-_36113502
|
0.538
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr2_+_102401580
|
0.536
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr10_-_105835525
|
0.535
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr4_+_35959638
|
0.532
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr1_+_87569889
|
0.530
|
|
|
|
|
chr2_+_27573206
|
0.524
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr19_+_54776398
|
0.522
|
NM_000951
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2
|
|
chr12_-_69434650
|
0.520
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr3_+_186529376
|
0.516
|
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr17_-_59817456
|
0.514
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr3_-_143230124
|
0.511
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr5_+_147562531
|
0.510
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr4_-_76817587
|
0.510
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr7_+_123036312
|
0.509
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr17_-_36074812
|
0.505
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr14_-_72563497
|
0.503
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr14_+_19761708
|
0.502
|
NM_001004480
|
OR11H6
|
olfactory receptor, family 11, subfamily H, member 6
|
|
chr16_+_54158084
|
0.499
|
NM_032330
|
CAPNS2
|
calpain, small subunit 2
|
|
chr1_+_61319976
|
0.498
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_559129
|
0.491
|
|
|
|
|
chr6_-_136889302
|
0.483
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_-_10453409
|
0.480
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr3_+_157877146
|
0.476
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr7_-_100077072
|
0.476
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr4_-_122304925
|
0.464
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr1_+_173111306
|
0.463
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr12_-_69434598
|
0.463
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_+_137701122
|
0.462
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr2_+_203207912
|
0.460
|
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr12_-_23995109
|
0.459
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_76817582
|
0.456
|
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr9_+_95886558
|
0.451
|
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr12_-_120725210
|
0.451
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr6_-_138862273
|
0.449
|
|
|
|
|
chr1_+_61697209
|
0.446
|
|
NFIA
|
nuclear factor I/A
|
|
chr13_-_98428244
|
0.446
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr3_-_15814678
|
0.446
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr7_-_96471544
|
0.445
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr8_-_110724594
|
0.444
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr10_-_116241572
|
0.443
|
|
|
|
|
chr6_-_130578110
|
0.436
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr8_+_102573837
|
0.436
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_8928508
|
0.433
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chr1_+_87569959
|
0.433
|
|
LMO4
|
LIM domain only 4
|
|
chr22_+_38213174
|
0.433
|
NM_001098270
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr2_+_108604111
|
0.431
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr21_+_38550739
|
0.427
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr1_-_150805852
|
0.422
|
NM_178435
|
LCE3E
|
late cornified envelope 3E
|
|
chr15_-_91078307
|
0.421
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr7_-_83662152
|
0.421
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr12_-_49708279
|
0.420
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr4_-_68678181
|
0.420
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chr16_-_3870121
|
0.414
|
NM_001079846
NM_004380
|
CREBBP
|
CREB binding protein
|
|
chr9_+_95886625
|
0.414
|
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr7_-_107558021
|
0.413
|
NM_007356
|
LAMB4
|
laminin, beta 4
|
|
chr1_-_57057956
|
0.413
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chr11_-_26550243
|
0.411
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr17_-_1359745
|
0.409
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr8_+_26206648
|
0.408
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr6_-_138862271
|
0.406
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr11_+_93940121
|
0.405
|
NM_152431
|
PIWIL4
|
piwi-like 4 (Drosophila)
|
|
chr16_+_24459173
|
0.403
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr6_+_57019269
|
0.402
|
NM_020931
|
KIAA1586
|
KIAA1586
|
|
chr21_-_30510113
|
0.402
|
NM_199328
|
CLDN8
|
claudin 8
|
|
chr15_+_41265429
|
0.401
|
NM_037370
|
CCNDBP1
|
cyclin D-type binding-protein 1
|
|
chr1_+_29113589
|
0.401
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr16_-_69876262
|
0.400
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr10_-_123346148
|
0.399
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_-_51660328
|
0.394
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr13_+_92677012
|
0.393
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr9_+_96807107
|
0.392
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr12_-_28016182
|
0.391
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr22_+_17498320
|
0.386
|
NM_053006
|
TSSK2
|
testis-specific serine kinase 2
|
|
chr22_+_21852551
|
0.386
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr2_+_233417348
|
0.383
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chrX_-_49852397
|
0.382
|
NM_003886
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr15_-_40089736
|
0.380
|
NM_001080490
|
PLA2G4E
|
phospholipase A2, group IVE
|
|
chr12_-_69317441
|
0.378
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr1_+_183970093
|
0.376
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr9_+_138810610
|
0.376
|
NM_001039374
|
KIAA1984
|
KIAA1984
|
|
chr19_-_14771947
|
0.376
|
NM_198944
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1
|
|
chr8_+_22266256
|
0.375
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr2_+_113452066
|
0.375
|
NM_019618
|
IL1F9
|
interleukin 1 family, member 9
|
|
chr12_-_49763405
|
0.375
|
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr1_-_199742589
|
0.374
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_+_142587973
|
0.374
|
|
|
|
|
chr9_+_102275310
|
0.372
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr4_+_77575276
|
0.370
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr5_+_133735004
|
0.369
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr7_-_142369519
|
0.368
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr11_+_73037382
|
0.368
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr3_-_14963943
|
0.367
|
|
|
|
|
chr5_+_34723420
|
0.366
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr14_-_54948290
|
0.365
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr9_+_102275455
|
0.364
|
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr7_-_13992351
|
0.363
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr3_-_129777618
|
0.363
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr7_-_107667728
|
0.362
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr22_-_34566367
|
0.361
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_151051070
|
0.360
|
NM_178349
|
LCE1B
|
late cornified envelope 1B
|
|
chr15_+_83467295
|
0.358
|
|
PDE8A
|
phosphodiesterase 8A
|
|
chr9_-_40782062
|
0.357
|
NM_033160
|
ZNF658
|
zinc finger protein 658
|
|
chr6_+_47732181
|
0.357
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr20_+_58064360
|
0.355
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr6_-_150253729
|
0.353
|
NM_139165
|
RAET1E
|
retinoic acid early transcript 1E
|
|
chr2_+_74914742
|
0.351
|
|
HK2
|
hexokinase 2
|
|
chr12_-_69434330
|
0.351
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr1_+_198975227
|
0.350
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|