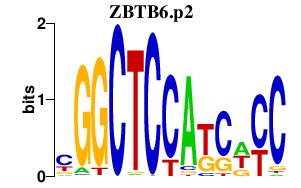
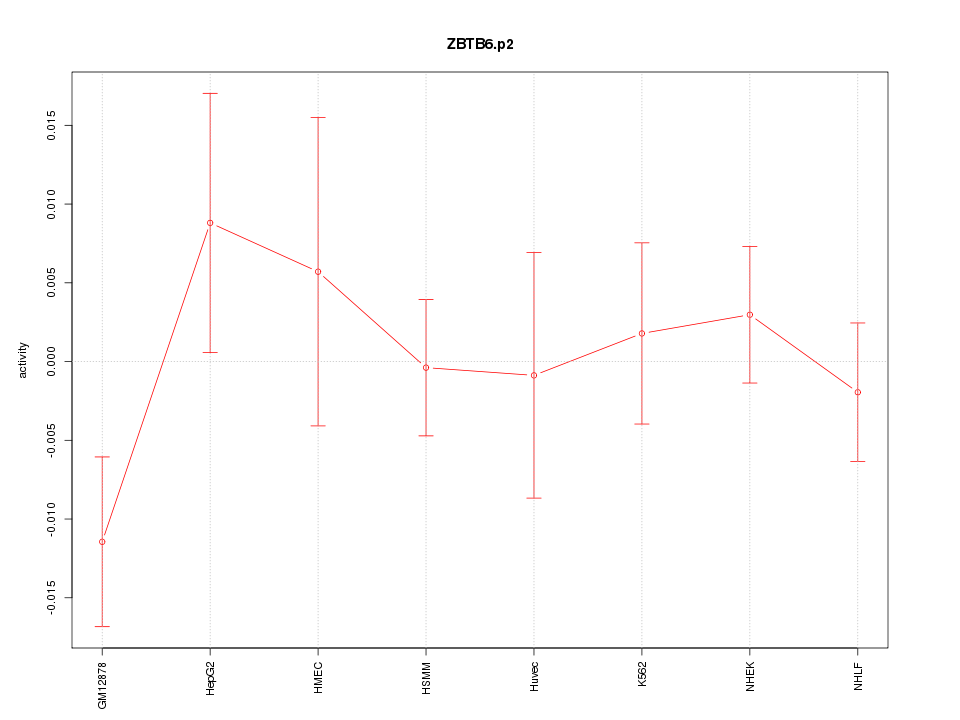
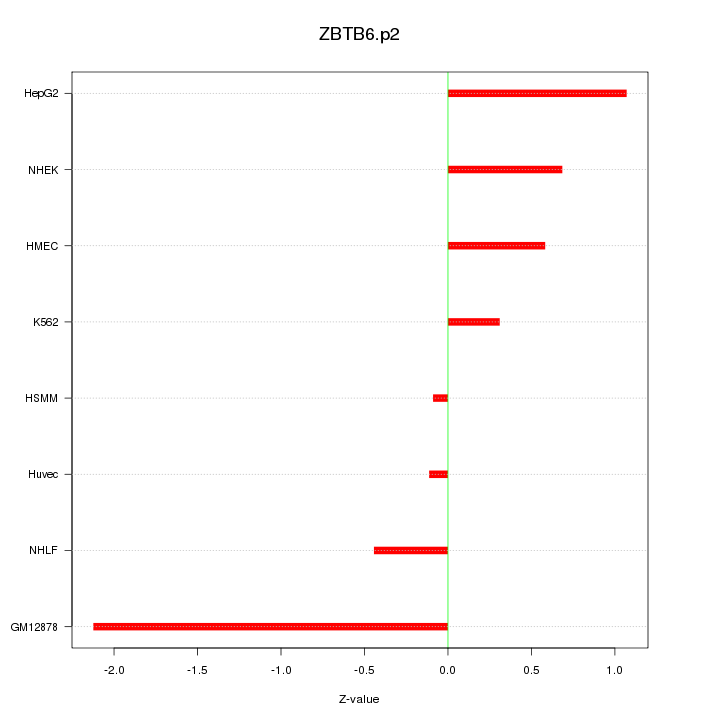
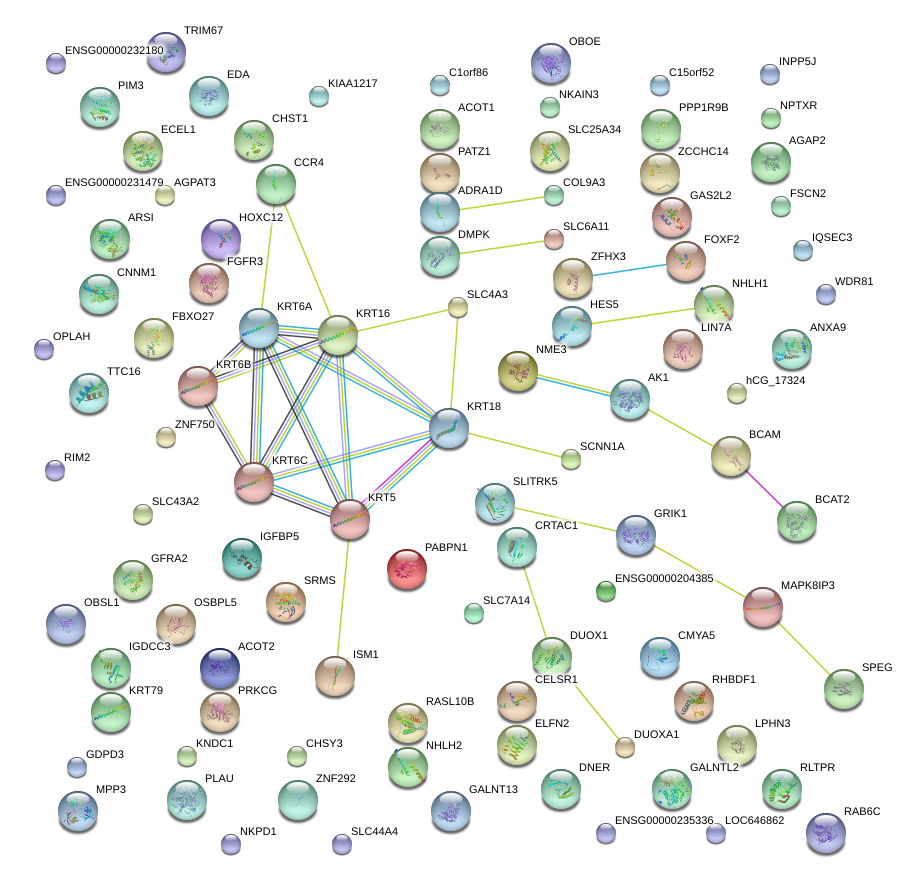
|
chr16_+_65018316
|
0.894
|
NM_001136106
NM_001178020
NM_001197224
NM_001197225
|
BEAN1
|
brain expressed, associated with NEDD4, 1
|
|
chr3_+_10832867
|
0.851
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr1_+_149221122
|
0.834
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr12_-_51200347
|
0.713
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr8_+_63324053
|
0.712
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr5_-_149662640
|
0.712
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr20_-_4177519
|
0.711
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr2_+_220017617
|
0.668
|
|
SPEG
|
SPEG complex locus
|
|
chr17_-_39265705
|
0.634
|
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr22_-_37569962
|
0.617
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr12_-_51153788
|
0.583
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr3_-_62835601
|
0.580
|
|
|
|
|
chr16_-_30032330
|
0.563
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr2_+_120018494
|
0.557
|
NM_001029996
|
PCDP1
|
primary ciliary dyskinesia protein 1
|
|
chr5_+_79021401
|
0.553
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr10_-_99780574
|
0.546
|
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr22_-_30071565
|
0.531
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr17_+_1580504
|
0.528
|
|
WDR81
|
WD repeat domain 81
|
|
chr19_-_50355247
|
0.522
|
NM_198478
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1
|
|
chr12_-_51173238
|
0.520
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr2_-_233060774
|
0.515
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr1_-_2129031
|
0.506
|
NM_001146310
|
C1orf86
|
chromosome 1 open reading frame 86
|
|
chr21_-_30234100
|
0.500
|
NM_000830
NM_175611
|
GRIK1
|
glutamate receptor, ionotropic, kainate 1
|
|
chr15_+_43209422
|
0.497
|
NM_017434
NM_175940
|
DUOX1
|
dual oxidase 1
|
|
chr21_+_44169706
|
0.493
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr4_+_61749540
|
0.468
|
|
LPHN3
|
latrophilin 3
|
|
chr17_+_77110011
|
0.465
|
NM_001077182
NM_012418
|
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus)
|
|
chr22_+_29848923
|
0.464
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr12_-_51132129
|
0.457
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr9_-_129679760
|
0.450
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr2_-_131838200
|
0.441
|
NM_001077637
|
LOC150786
|
RAB6C-like
|
|
chr12_+_52634950
|
0.431
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr15_-_38420414
|
0.430
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr17_-_45582872
|
0.417
|
NM_032595
|
PPP1R9B
|
protein phosphatase 1, regulatory (inhibitor) subunit 9B
|
|
chr8_-_21702273
|
0.413
|
NM_001165038
NM_001165039
NM_001495
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr17_-_39265965
|
0.412
|
NM_001932
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3)
|
|
chr12_+_46133
|
0.410
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr16_-_86082818
|
0.410
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr17_+_31082791
|
0.395
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr20_+_60918836
|
0.390
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr10_+_101079099
|
0.388
|
|
CNNM1
|
cyclin M1
|
|
chr17_-_78391178
|
0.384
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr2_-_217267742
|
0.382
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr2_-_230287442
|
0.379
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr1_-_226530350
|
0.378
|
|
|
|
|
chr3_-_171786468
|
0.376
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr10_+_75343352
|
0.367
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr15_-_63457378
|
0.365
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr20_-_61649259
|
0.363
|
NM_080823
|
SRMS
|
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites
|
|
chr1_-_48235148
|
0.362
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chrX_+_68752635
|
0.360
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr1_+_15935395
|
0.360
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr16_-_71650034
|
0.360
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr4_+_1764802
|
0.354
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr2_-_220144254
|
0.350
|
|
OBSL1
|
obscurin-like 1
|
|
chr12_-_56418295
|
0.347
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr17_-_31104009
|
0.346
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr22_+_48740432
|
0.345
|
|
PIM3
|
pim-3 oncogene
|
|
chr20_+_13150417
|
0.343
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr10_+_101078784
|
0.339
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr17_-_37022465
|
0.338
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr10_+_24023680
|
0.336
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr16_-_62573
|
0.334
|
NM_022450
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila)
|
|
chr16_+_66244314
|
0.326
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr22_-_45311730
|
0.325
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr14_+_22860511
|
0.323
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr19_-_50977586
|
0.322
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr1_+_229365296
|
0.321
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr19_+_50004134
|
0.319
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr6_+_87921980
|
0.315
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr11_-_3104413
|
0.314
|
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr19_+_63599268
|
0.308
|
NM_001195135
|
LOC646862
|
hypothetical protein LOC646862
|
|
chr22_-_30071761
|
0.308
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr6_-_31954795
|
0.306
|
NM_001178044
NM_025257
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr22_-_36153449
|
0.303
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr1_+_116181927
|
0.303
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr19_-_44214972
|
0.299
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr13_+_87122778
|
0.297
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr2_+_220033777
|
0.297
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr12_-_79855580
|
0.295
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr9_+_129518163
|
0.294
|
NM_144965
|
TTC16
|
tetratricopeptide repeat domain 16
|
|
chr10_-_99780336
|
0.294
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr12_+_51628921
|
0.294
|
NM_199187
NM_000224
|
KRT18
|
keratin 18
|
|
chr6_+_1335067
|
0.294
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr15_-_43209348
|
0.293
|
NM_144565
|
DUOXA1
|
dual oxidase maturation factor 1
|
|
chr16_-_1761710
|
0.290
|
NM_002513
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr8_-_145187553
|
0.290
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr19_+_59077241
|
0.287
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr16_+_1696162
|
0.284
|
NM_001040439
NM_015133
|
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr5_+_129268352
|
0.283
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr8_+_104582151
|
0.281
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr6_-_31954745
|
0.280
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr2_+_220200530
|
0.280
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr12_-_6354650
|
0.279
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr6_-_31954730
|
0.278
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr10_+_134823925
|
0.277
|
NM_152643
|
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1
|
|
chr7_-_20791907
|
0.277
|
|
|
|
|
chr11_-_45643711
|
0.275
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr2_+_154436670
|
0.274
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr1_-_2451543
|
0.274
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr14_+_73073570
|
0.273
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr17_-_1478293
|
0.273
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr5_+_167889123
|
0.270
|
|
|
|
|
chr20_+_2581177
|
0.269
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr19_-_54267932
|
0.268
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr10_+_47875229
|
0.268
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr2_-_219882302
|
0.267
|
NM_002846
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr6_-_136889302
|
0.266
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_+_20751518
|
0.266
|
NM_207334
|
FAM43B
|
family with sequence similarity 43, member B
|
|
chr19_+_39664706
|
0.265
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr20_-_9767406
|
0.262
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr9_-_129869211
|
0.261
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr14_+_22860547
|
0.259
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr2_+_223244606
|
0.255
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr12_-_6354818
|
0.254
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chrX_-_50230329
|
0.253
|
NM_001013742
|
DGKK
|
diacylglycerol kinase, kappa
|
|
chr17_-_40348378
|
0.252
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr2_-_18634257
|
0.252
|
NM_001002006
NM_033253
|
NT5C1B-RDH14
NT5C1B
|
NT5C1B-RDH14 readthrough
5'-nucleotidase, cytosolic IB
|
|
chr14_-_22974707
|
0.251
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr22_-_37569679
|
0.250
|
|
|
|
|
chr15_-_28472854
|
0.249
|
NM_139320
NM_148911
|
CHRFAM7A
|
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion
|
|
chr16_-_30288988
|
0.249
|
NM_015527
|
TBC1D10B
|
TBC1 domain family, member 10B
|
|
chr11_+_20647692
|
0.248
|
NM_006157
NM_201551
|
NELL1
|
NEL-like 1 (chicken)
|
|
chr3_+_161426322
|
0.248
|
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr20_-_49612745
|
0.246
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr8_+_145700272
|
0.246
|
NM_005309
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase)
|
|
chr3_+_197427779
|
0.244
|
NM_152672
|
OSTalpha
|
organic solute transporter alpha
|
|
chr13_+_92677012
|
0.241
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr14_+_104261578
|
0.241
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr3_+_182912769
|
0.238
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr14_-_37134191
|
0.237
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr16_-_29532538
|
0.237
|
|
|
|
|
chr3_-_38046034
|
0.237
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr14_-_65550379
|
0.237
|
|
|
|
|
chr2_+_27225362
|
0.235
|
NM_175769
|
TCF23
|
transcription factor 23
|
|
chr13_-_37341859
|
0.233
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr18_-_24011349
|
0.233
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr11_+_82545784
|
0.233
|
NM_015885
|
PCF11
|
PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae)
|
|
chr11_+_125781157
|
0.232
|
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr1_+_109594163
|
0.230
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr9_-_103540682
|
0.229
|
NM_133445
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr6_+_108593907
|
0.229
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr17_+_2627099
|
0.229
|
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr3_+_142433361
|
0.229
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr13_+_27392136
|
0.228
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr9_-_103540038
|
0.228
|
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr17_+_4434582
|
0.228
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr17_-_39757725
|
0.227
|
NM_001143780
NM_016016
|
SLC25A39
|
solute carrier family 25, member 39
|
|
chr11_+_72203111
|
0.227
|
|
ATG16L2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae)
|
|
chr10_+_30762862
|
0.225
|
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr4_+_134292630
|
0.225
|
|
PCDH10
|
protocadherin 10
|
|
chr3_-_13896618
|
0.225
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr19_-_50977485
|
0.224
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr16_+_86542449
|
0.221
|
NM_001173539
NM_001173540
NM_001173541
NM_017869
NM_079837
|
BANP
|
BTG3 associated nuclear protein
|
|
chr10_-_121292139
|
0.220
|
NM_001005339
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr10_+_81882409
|
0.220
|
|
PLAC9
|
placenta-specific 9
|
|
chr6_-_37773609
|
0.220
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr11_+_116575249
|
0.220
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr8_-_37943143
|
0.220
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr6_-_88932485
|
0.220
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr12_-_1573591
|
0.219
|
NM_152441
|
FBXL14
|
F-box and leucine-rich repeat protein 14
|
|
chr19_-_44214707
|
0.218
|
|
|
|
|
chr22_+_23149855
|
0.217
|
|
ADORA2A
|
adenosine A2a receptor
|
|
chr22_-_19122047
|
0.217
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr1_-_26245031
|
0.214
|
NM_001004434
NM_032513
|
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2
|
|
chr10_+_30763056
|
0.214
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr17_+_40681060
|
0.214
|
|
LOC100133991
|
similar to hCG1995169
|
|
chr6_-_2916980
|
0.214
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr18_+_2645885
|
0.213
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr3_-_62836052
|
0.212
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr16_-_21439195
|
0.212
|
|
SLC7A5P2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 pseudogene 2
|
|
chr20_-_24986501
|
0.211
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr19_+_45811316
|
0.211
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr12_-_53059234
|
0.210
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr4_-_66218777
|
0.209
|
|
EPHA5
|
EPH receptor A5
|
|
chr19_-_18409971
|
0.209
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr14_+_23937766
|
0.209
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr20_-_3166834
|
0.208
|
NM_001174090
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr17_-_70401299
|
0.208
|
NM_178128
|
FADS6
|
fatty acid desaturase domain family, member 6
|
|
chr16_-_1761504
|
0.207
|
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr17_-_6675731
|
0.207
|
NM_053285
|
TEKT1
|
tektin 1
|
|
chr11_+_62136788
|
0.207
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr1_-_218168137
|
0.206
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr11_-_65443015
|
0.206
|
NM_001135635
NM_031450
|
C11orf68
|
chromosome 11 open reading frame 68
|
|
chr1_-_221604023
|
0.206
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr12_-_79855618
|
0.206
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr10_+_72642274
|
0.206
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr16_+_29735208
|
0.206
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr1_+_15957832
|
0.205
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr3_+_161425946
|
0.204
|
NM_001168214
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr1_+_28071640
|
0.203
|
NM_001039477
NM_001105556
NM_004848
|
C1orf38
|
chromosome 1 open reading frame 38
|
|
chr2_-_72224696
|
0.203
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr10_+_81882477
|
0.202
|
|
PLAC9
|
placenta-specific 9
|
|
chr1_+_5975345
|
0.202
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr16_-_4105703
|
0.202
|
|
ADCY9
|
adenylate cyclase 9
|
|
chr17_-_45427565
|
0.202
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr10_+_116843108
|
0.201
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr1_+_237616457
|
0.201
|
|
CHRM3
|
cholinergic receptor, muscarinic 3
|
|
chr20_+_34357604
|
0.201
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr22_-_30072329
|
0.198
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|