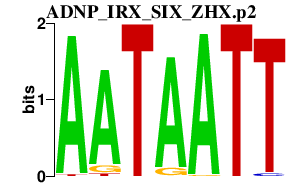
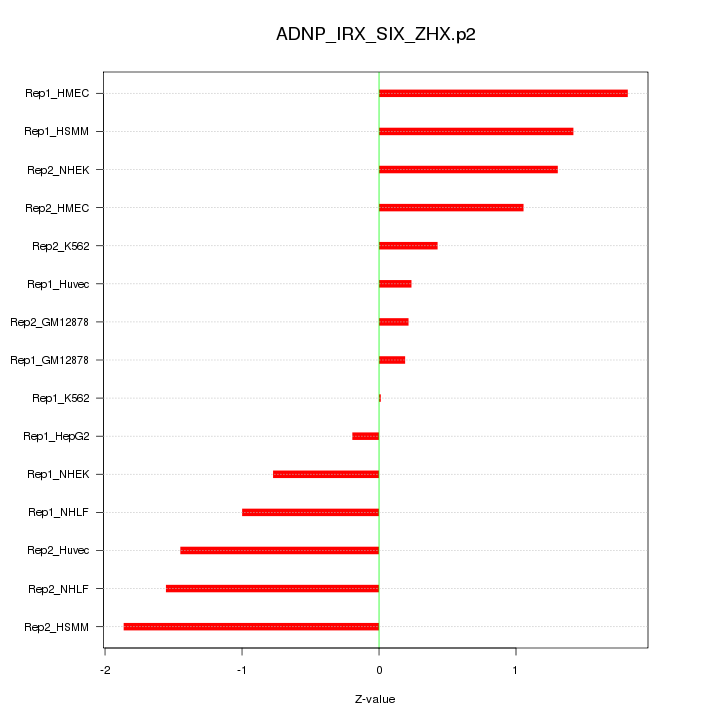
|
chr3_-_57209319
|
1.642
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr8_-_72431274
|
1.555
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chrX_-_15593074
|
1.315
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr11_-_83071084
|
1.173
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr5_+_147562531
|
1.086
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr11_-_123353607
|
1.064
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr1_-_108032645
|
0.973
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr3_-_110155237
|
0.945
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr18_-_59480082
|
0.917
|
NM_006919
|
SERPINB3
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr5_+_73145098
|
0.911
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr14_-_80934523
|
0.901
|
NM_033104
|
STON2
|
stonin 2
|
|
chr5_+_60969392
|
0.899
|
NM_173667
|
FLJ37543
|
hypothetical protein FLJ37543
|
|
chr18_-_26996742
|
0.891
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr11_-_104515670
|
0.874
|
NM_021571
|
CARD18
|
caspase recruitment domain family, member 18
|
|
chrX_-_137649180
|
0.849
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr6_-_52060327
|
0.848
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr3_-_55489973
|
0.840
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr1_+_556968
|
0.836
|
|
|
|
|
chr9_-_14076901
|
0.835
|
|
NFIB
|
nuclear factor I/B
|
|
chr10_+_124310170
|
0.823
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr15_+_91244563
|
0.817
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr12_-_51200347
|
0.812
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr8_-_17797127
|
0.785
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr11_-_5946299
|
0.771
|
NM_001146033
|
OR56A5
|
olfactory receptor, family 56, subfamily A, member 5
|
|
chr9_-_99740243
|
0.769
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr11_-_118405288
|
0.759
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr20_+_58064360
|
0.709
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr12_-_10983072
|
0.703
|
NM_023922
|
TAS2R14
|
taste receptor, type 2, member 14
|
|
chr18_+_19826734
|
0.693
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr4_-_70760381
|
0.684
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr3_-_58175437
|
0.681
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr8_-_86477585
|
0.677
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr2_+_96269703
|
0.651
|
NM_001037228
|
LOC285033
|
hypothetical protein LOC285033
|
|
chrX_-_18600108
|
0.648
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr6_+_167445284
|
0.641
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr21_-_30460841
|
0.634
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr20_+_58004811
|
0.624
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chr11_+_34599227
|
0.597
|
|
EHF
|
ets homologous factor
|
|
chr11_+_34599163
|
0.594
|
NM_012153
|
EHF
|
ets homologous factor
|
|
chr11_-_16454493
|
0.590
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_-_40173949
|
0.572
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr4_-_77147625
|
0.553
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr5_-_22889487
|
0.553
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr14_-_64479198
|
0.553
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr11_-_26550243
|
0.552
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr12_-_10497195
|
0.550
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr7_-_141293185
|
0.549
|
NM_013252
|
CLEC5A
|
C-type lectin domain family 5, member A
|
|
chr2_+_102401580
|
0.547
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr18_+_27152049
|
0.546
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr1_+_165565006
|
0.545
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr2_-_99238001
|
0.544
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr7_-_141604323
|
0.542
|
NM_001001317
|
TRYX3
|
trypsin X3
|
|
chr22_-_34566367
|
0.537
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_32892705
|
0.537
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr19_-_51236113
|
0.536
|
NM_001002923
|
IGFL4
|
IGF-like family member 4
|
|
chr1_+_200246312
|
0.535
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr1_+_78243181
|
0.533
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr7_-_93027057
|
0.529
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chrX_+_85404626
|
0.529
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr1_+_200246370
|
0.523
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr3_+_88270951
|
0.521
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr8_+_12847521
|
0.520
|
NM_020844
|
KIAA1456
|
KIAA1456
|
|
chr3_+_187840842
|
0.520
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr9_-_205892
|
0.516
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr11_+_10433241
|
0.513
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_+_99465814
|
0.508
|
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr11_+_65027721
|
0.507
|
|
|
|
|
chr9_+_27514311
|
0.503
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr15_-_64108718
|
0.501
|
|
|
|
|
chr3_+_115099007
|
0.501
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr12_-_51173238
|
0.498
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr4_-_91979113
|
0.495
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr1_+_165564904
|
0.492
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr5_-_168204430
|
0.479
|
|
|
|
|
chr13_+_107753764
|
0.476
|
|
|
|
|
chr16_+_2593351
|
0.475
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr17_-_37022465
|
0.475
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr2_-_207291364
|
0.474
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr3_+_123256902
|
0.471
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr15_+_91244422
|
0.469
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr7_+_128802719
|
0.465
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_+_117452995
|
0.461
|
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr7_-_25234582
|
0.457
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr6_+_21701942
|
0.455
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr11_+_120478584
|
0.446
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr7_-_16811131
|
0.445
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr5_+_141326620
|
0.439
|
NM_183399
|
RNF14
|
ring finger protein 14
|
|
chr5_+_156628929
|
0.438
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr12_+_28301399
|
0.437
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr11_-_128399218
|
0.436
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr20_-_33658823
|
0.434
|
|
FER1L4
|
fer-1-like 4 (C. elegans) pseudogene
|
|
chr2_-_29150630
|
0.429
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr13_-_85271329
|
0.429
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr18_+_30812205
|
0.426
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr17_-_64752550
|
0.425
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr21_+_38550739
|
0.425
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr4_+_70831387
|
0.421
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chr14_-_68424653
|
0.421
|
|
ACTN1
|
actinin, alpha 1
|
|
chr2_-_98646367
|
0.420
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr1_+_158975662
|
0.414
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chr1_+_86662356
|
0.411
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr2_+_161981138
|
0.410
|
|
TBR1
|
T-box, brain, 1
|
|
chr17_-_25685175
|
0.410
|
NM_206832
|
TMIGD1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr20_-_43601542
|
0.405
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr10_+_63331401
|
0.405
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr2_-_96927557
|
0.403
|
NM_016490
|
FAM178B
|
family with sequence similarity 178, member B
|
|
chr5_+_67622242
|
0.402
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr5_-_22889192
|
0.399
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr1_-_205272619
|
0.398
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr3_+_137224266
|
0.397
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_-_98021144
|
0.396
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr12_+_52696908
|
0.392
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr4_-_77547481
|
0.392
|
NM_001042784
|
CCDC158
|
coiled-coil domain containing 158
|
|
chr5_-_148738980
|
0.392
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr4_+_106693225
|
0.389
|
NM_017700
|
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38
|
|
chr11_-_64484184
|
0.388
|
NM_001037225
|
C11orf85
|
chromosome 11 open reading frame 85
|
|
chr8_+_77756061
|
0.385
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr17_-_36347197
|
0.384
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_-_138862273
|
0.382
|
|
|
|
|
chr2_+_234266250
|
0.382
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr2_+_11591692
|
0.382
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chr5_-_1935764
|
0.382
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chrX_-_15198093
|
0.380
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr6_+_29534109
|
0.379
|
NM_030883
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1
|
|
chr7_-_92693716
|
0.378
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr1_-_114215768
|
0.377
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr15_+_56217686
|
0.374
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr1_-_151352587
|
0.373
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr11_+_123600553
|
0.373
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr18_+_59726183
|
0.372
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr5_+_59819761
|
0.371
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr3_+_112743615
|
0.370
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chrX_+_110074124
|
0.368
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr2_-_227952249
|
0.367
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr12_+_52697160
|
0.364
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr4_-_72868598
|
0.364
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr4_-_153493133
|
0.364
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr11_+_20577521
|
0.363
|
NM_004211
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5
|
|
chr4_+_120029426
|
0.359
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chrX_+_99726445
|
0.355
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr3_+_46370228
|
0.354
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr8_+_77756077
|
0.349
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr1_+_86785346
|
0.347
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr13_-_83354425
|
0.345
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr12_-_51153788
|
0.345
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr12_-_51476047
|
0.345
|
NM_057088
|
KRT3
|
keratin 3
|
|
chr12_-_55614311
|
0.344
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr1_+_35623688
|
0.343
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr7_-_16471817
|
0.342
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr5_-_134290789
|
0.342
|
|
|
|
|
chr12_-_16321885
|
0.340
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr13_-_83354474
|
0.339
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr2_-_20390601
|
0.338
|
NM_015317
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr9_+_106496480
|
0.337
|
NM_001004484
|
OR13D1
|
olfactory receptor, family 13, subfamily D, member 1
|
|
chr10_+_68355797
|
0.337
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chrX_+_107174855
|
0.336
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr4_+_88973144
|
0.336
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chrX_-_15198509
|
0.333
|
NM_001168531
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr15_+_38319326
|
0.331
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr1_+_205068844
|
0.331
|
NM_013371
|
IL19
|
interleukin 19
|
|
chr4_-_74704964
|
0.329
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr12_-_14024187
|
0.328
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr8_-_72436519
|
0.328
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_+_161981046
|
0.327
|
|
TBR1
|
T-box, brain, 1
|
|
chr3_-_156904690
|
0.327
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr13_+_77007809
|
0.325
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr22_-_34566276
|
0.325
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr10_+_49279647
|
0.323
|
NM_002750
NM_139046
NM_139047
NM_139049
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr7_-_135263737
|
0.323
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr2_-_1905508
|
0.323
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr17_-_36192311
|
0.322
|
NM_181537
|
KRT27
|
keratin 27
|
|
chr10_-_85975263
|
0.320
|
NM_001017924
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2
|
|
chr10_+_4995453
|
0.318
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr1_+_234624266
|
0.317
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr1_-_163680941
|
0.317
|
NM_006917
|
RXRG
|
retinoid X receptor, gamma
|
|
chr20_+_57966865
|
0.316
|
NM_177980
|
CDH26
|
cadherin 26
|
|
chr12_-_101036407
|
0.316
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chrX_+_70682204
|
0.315
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr4_-_54152385
|
0.312
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr4_-_87989291
|
0.312
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr6_+_7524691
|
0.312
|
|
DSP
|
desmoplakin
|
|
chr14_-_93493519
|
0.310
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr3_+_176059763
|
0.308
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr12_+_103677388
|
0.306
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr4_+_109033868
|
0.305
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr4_+_47181987
|
0.304
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chr11_+_117452936
|
0.303
|
NM_001083947
NM_001173551
NM_001173552
NM_019894
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chrX_+_37750778
|
0.302
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr19_+_12995782
|
0.301
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_+_130745580
|
0.301
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr4_-_122304925
|
0.297
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr3_+_112743729
|
0.297
|
|
CD96
|
CD96 molecule
|
|
chr19_+_40632456
|
0.296
|
NM_005306
|
FFAR2
|
free fatty acid receptor 2
|
|
chr10_+_70172257
|
0.295
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr2_-_69033515
|
0.294
|
NM_182536
|
GKN2
|
gastrokine 2
|
|
chr2_+_161980865
|
0.293
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr15_-_74091817
|
0.293
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr3_-_56673113
|
0.293
|
NM_015224
|
C3orf63
|
chromosome 3 open reading frame 63
|
|
chr4_+_68995761
|
0.292
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr4_+_76700281
|
0.291
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|