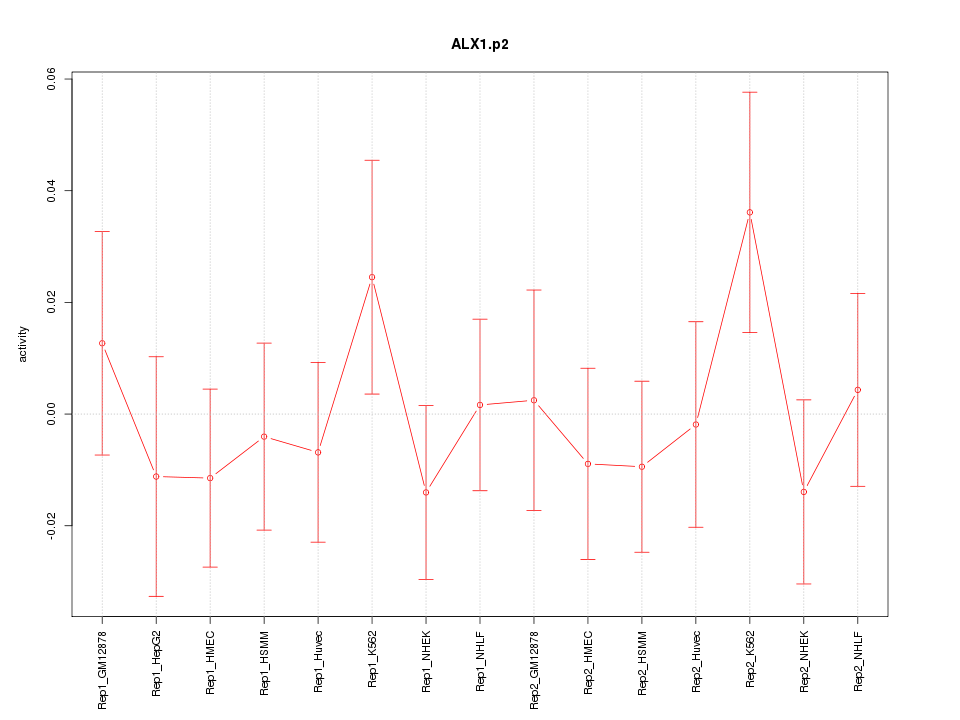
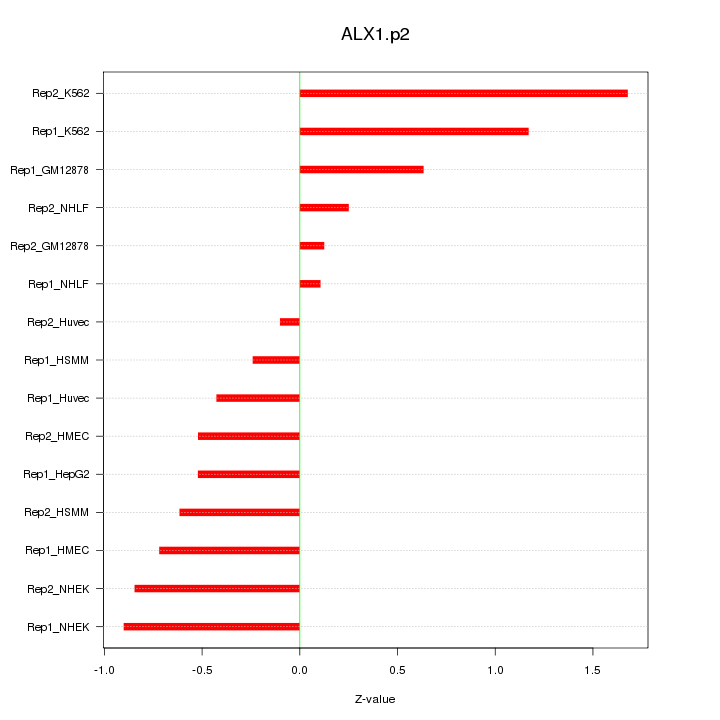
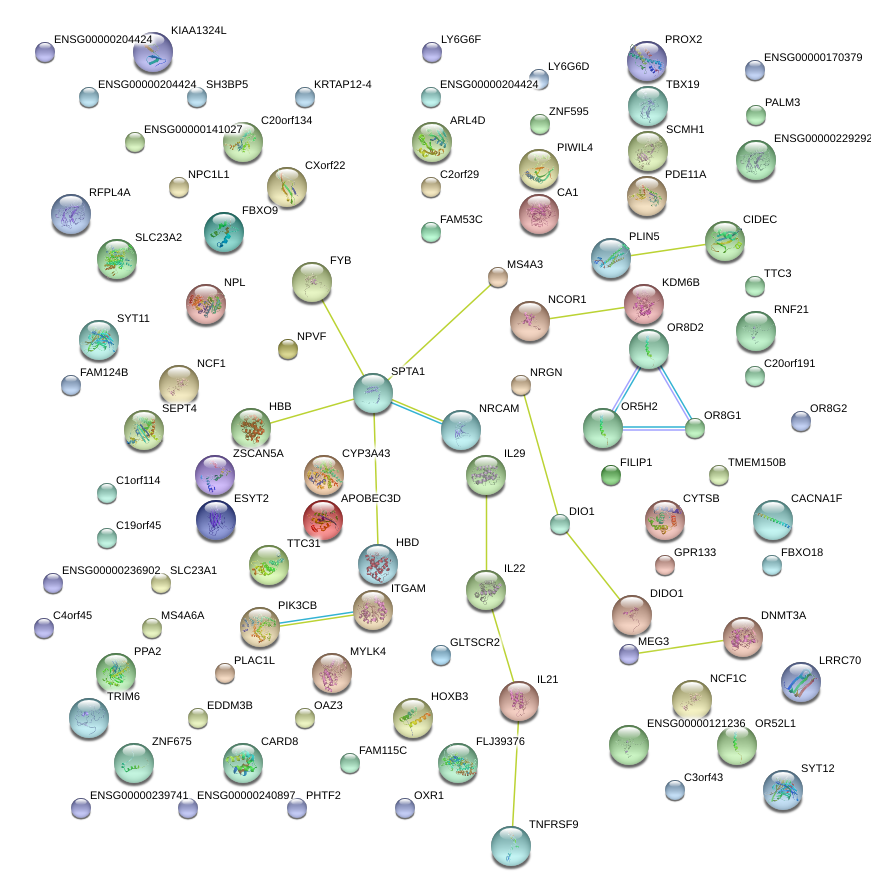
|
chr11_-_5212168
|
3.257
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr7_-_107667728
|
2.061
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr1_+_166516901
|
2.056
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr12_+_130004404
|
1.660
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr8_-_86477585
|
1.526
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr1_-_41399632
|
1.449
|
NM_001031694
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr11_+_59580630
|
1.438
|
NM_001031666
NM_001031809
NM_006138
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr17_+_19917420
|
1.399
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr7_-_44547382
|
1.137
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr19_-_14030970
|
1.121
|
NM_001145028
|
PALM3
|
paralemmin 3
|
|
chr4_+_43185
|
1.107
|
NM_182524
|
ZNF595
ZNF718
|
zinc finger protein 595
zinc finger protein 718
|
|
chr19_-_53444733
|
1.028
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr6_-_2696152
|
0.999
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr1_+_181028016
|
0.995
|
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr7_+_77307382
|
0.946
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr5_-_138746840
|
0.865
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr10_+_5976354
|
0.848
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr1_+_154120170
|
0.833
|
|
SYT11
|
synaptotagmin XI
|
|
chr6_+_53043729
|
0.826
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr3_-_197726587
|
0.819
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr11_-_59707074
|
0.818
|
NM_022349
NM_152851
NM_152852
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A
|
|
chr7_+_73826244
|
0.811
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr11_-_123695302
|
0.795
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr2_-_224974954
|
0.782
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr7_-_25234582
|
0.780
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr1_+_559618
|
0.776
|
|
|
|
|
chr17_-_16038607
|
0.775
|
NM_001190438
NM_001190440
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr11_+_123600553
|
0.759
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr17_-_44006808
|
0.753
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr14_+_20306425
|
0.742
|
NM_022360
|
EDDM3B
|
epididymal protein 3B
|
|
chr1_+_150005754
|
0.738
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr7_-_158304062
|
0.711
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr6_+_31791063
|
0.699
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr19_+_7468444
|
0.686
|
NM_198534
|
C19orf45
|
chromosome 19 open reading frame 45
|
|
chr11_+_123625632
|
0.659
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr5_+_137701122
|
0.644
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr14_-_74400288
|
0.644
|
NM_001080408
|
PROX2
|
prospero homeobox 2
|
|
chr4_-_123761615
|
0.637
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr7_-_86433139
|
0.629
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr11_+_5610060
|
0.621
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr1_+_559129
|
0.621
|
|
|
|
|
chr5_-_39306465
|
0.618
|
|
FYB
|
FYN binding protein
|
|
chr19_-_60528480
|
0.607
|
NM_001085488
|
TMEM150B
|
transmembrane protein 150B
|
|
chrX_-_48976714
|
0.601
|
NM_005183
|
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit
|
|
chr8_+_107807415
|
0.598
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr1_-_7923464
|
0.595
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr3_-_15357878
|
0.589
|
NM_001018009
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr14_+_100365390
|
0.576
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr6_-_76259976
|
0.570
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr16_+_31178828
|
0.566
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr19_+_44478804
|
0.559
|
NM_172140
|
IL29
|
interleukin 29 (interferon, lambda 1)
|
|
chr19_+_60962318
|
0.544
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr21_-_44899003
|
0.529
|
NM_198698
|
KRTAP12-4
|
keratin associated protein 12-4
|
|
chr7_+_143043871
|
0.520
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr10_-_42183441
|
0.517
|
|
LOC283027
|
hypothetical protein LOC283027
|
|
chr11_+_93940121
|
0.514
|
NM_152431
|
PIWIL4
|
piwi-like 4 (Drosophila)
|
|
chr16_+_31178778
|
0.509
|
NM_000632
NM_001145808
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr21_+_37377116
|
0.508
|
NM_003316
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr19_-_4486202
|
0.508
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr1_+_54132447
|
0.504
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr16_+_31178812
|
0.502
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr4_-_160175782
|
0.499
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr3_-_9895633
|
0.494
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr20_+_31717964
|
0.486
|
NM_001024675
|
C20orf134
|
chromosome 20 open reading frame 134
|
|
chr2_-_74235860
|
0.486
|
|
|
|
|
chr5_+_61910318
|
0.480
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr1_-_156923111
|
0.480
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr22_-_20667131
|
0.479
|
NM_003935
|
TOP3B
|
topoisomerase (DNA) III beta
|
|
chr2_-_178681311
|
0.479
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr3_-_139960874
|
0.478
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr2_-_25320299
|
0.474
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr19_-_23661779
|
0.473
|
NM_138330
|
ZNF675
|
zinc finger protein 675
|
|
chr11_+_59564323
|
0.467
|
NM_173801
|
PLAC1L
|
placenta-specific 1-like
|
|
chr19_-_61431470
|
0.463
|
NM_024303
|
ZSCAN5A
|
zinc finger and SCAN domain containing 5A
|
|
chr7_+_99263571
|
0.461
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr8_-_110730183
|
0.459
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr2_-_127817143
|
0.455
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr10_+_35524060
|
0.446
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_67404756
|
0.445
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr1_-_201044168
|
0.444
|
NM_006618
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr19_-_56564068
|
0.443
|
NM_152353
|
CLDND2
|
claudin domain containing 2
|
|
chr3_-_25658829
|
0.440
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chrX_-_11194015
|
0.438
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr9_+_89687560
|
0.431
|
NM_178828
|
C9orf79
|
chromosome 9 open reading frame 79
|
|
chr19_+_21116666
|
0.429
|
NM_133473
|
ZNF431
|
zinc finger protein 431
|
|
chr1_-_51757490
|
0.429
|
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr5_-_37220702
|
0.427
|
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr9_+_99807257
|
0.427
|
|
|
|
|
chr7_+_101810018
|
0.425
|
|
|
|
|
chr5_+_76470829
|
0.419
|
|
|
|
|
chr6_-_25982418
|
0.419
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr1_-_201044137
|
0.417
|
|
KDM5B
|
lysine (K)-specific demethylase 5B
|
|
chr1_+_156991133
|
0.414
|
NM_001005184
|
OR6K6
|
olfactory receptor, family 6, subfamily K, member 6
|
|
chrX_-_153716130
|
0.407
|
NM_001162936
|
LOC100132963
|
hypothetical protein LOC100132963
|
|
chr6_+_27215039
|
0.403
|
NM_003495
|
HIST1H4I
|
histone cluster 1, H4i
|
|
chr12_+_53177806
|
0.395
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr12_+_53177789
|
0.393
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr1_-_180908639
|
0.392
|
NM_001102450
NM_033345
|
RGS8
|
regulator of G-protein signaling 8
|
|
chr12_+_9033403
|
0.390
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr11_+_120478584
|
0.382
|
NM_005422
|
TECTA
|
tectorin alpha
|
|
chr12_+_53177758
|
0.378
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_+_59875623
|
0.378
|
NM_001085423
|
C17orf60
|
chromosome 17 open reading frame 60
|
|
chr19_+_21480229
|
0.372
|
NM_001001415
|
ZNF429
|
zinc finger protein 429
|
|
chr14_-_105542441
|
0.371
|
|
IGLJ3
IGHA1
IGHG1
IGHM
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr1_+_200584421
|
0.368
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr1_+_234624266
|
0.356
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr1_-_67039518
|
0.343
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr3_+_123817151
|
0.340
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr19_+_53450743
|
0.338
|
|
|
|
|
chr19_+_60053709
|
0.336
|
NM_006737
|
KIR3DL2
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 2
|
|
chr1_+_242582559
|
0.335
|
NM_001012970
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr1_+_190394214
|
0.322
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr7_+_37689903
|
0.321
|
|
|
|
|
chr9_-_122852356
|
0.320
|
NM_001735
|
C5
|
complement component 5
|
|
chr1_-_89414310
|
0.317
|
NM_207398
|
GBP7
|
guanylate binding protein 7
|
|
chr1_+_117345894
|
0.317
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr2_+_169637255
|
0.316
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr18_+_8708448
|
0.315
|
|
|
|
|
chr3_-_87408301
|
0.313
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr2_-_162717159
|
0.311
|
NM_002054
|
GCG
|
glucagon
|
|
chr11_+_46255737
|
0.310
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr22_-_22252376
|
0.307
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr7_+_63305015
|
0.302
|
NM_001159524
|
ZNF735
|
zinc finger protein 735
|
|
chr18_-_50134940
|
0.300
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr4_-_164754106
|
0.299
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr14_+_19735334
|
0.299
|
NM_001005503
|
OR11G2
|
olfactory receptor, family 11, subfamily G, member 2
|
|
chr7_-_107930170
|
0.293
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr20_+_56912146
|
0.287
|
|
GNAS
|
GNAS complex locus
|
|
chr4_+_68107039
|
0.286
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr3_+_122769467
|
0.286
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr2_-_68952006
|
0.286
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr12_+_9925354
|
0.282
|
NM_001190765
|
KLRF2
|
killer cell lectin-like receptor subfamily F, member 2
|
|
chr2_-_88946767
|
0.282
|
|
|
|
|
chr17_+_69781980
|
0.278
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chrX_-_151370325
|
0.274
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr19_+_59941785
|
0.273
|
NM_015868
|
KIR2DS2
KIR2DL3
|
killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 2
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 3
|
|
chr20_+_51235228
|
0.272
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr11_-_16387001
|
0.269
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_+_113351006
|
0.267
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr7_-_44834702
|
0.265
|
|
H2AFV
|
H2A histone family, member V
|
|
chr5_-_176258920
|
0.263
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr11_-_64459910
|
0.260
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr1_-_157014048
|
0.258
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chr14_-_22574193
|
0.258
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr11_-_10546654
|
0.257
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr13_-_98757634
|
0.257
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr19_+_40824486
|
0.252
|
NM_014209
|
ETV2
|
ets variant 2
|
|
chr21_+_37714470
|
0.250
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr11_+_123639909
|
0.249
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr19_+_61039755
|
0.249
|
NM_134444
|
NLRP4
|
NLR family, pyrin domain containing 4
|
|
chr8_-_37876129
|
0.247
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr9_-_27287106
|
0.247
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr19_+_7410573
|
0.245
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr5_-_132228375
|
0.243
|
NM_005260
|
GDF9
|
growth differentiation factor 9
|
|
chr4_-_72868598
|
0.241
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr3_-_69120188
|
0.241
|
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr10_-_50066386
|
0.240
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr16_-_79811475
|
0.240
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr1_-_20014180
|
0.240
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chrX_+_108665665
|
0.236
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr19_-_21438525
|
0.235
|
|
|
|
|
chr7_+_142590633
|
0.235
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chr4_+_55956520
|
0.235
|
|
|
|
|
chr7_+_115650240
|
0.234
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr2_-_63518589
|
0.234
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr22_-_20667165
|
0.233
|
|
TOP3B
|
topoisomerase (DNA) III beta
|
|
chr12_-_69434598
|
0.233
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr19_-_44518516
|
0.232
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr20_+_31713750
|
0.231
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr10_+_43187095
|
0.231
|
NM_001184963
NM_173160
|
FXYD4
|
FXYD domain containing ion transport regulator 4
|
|
chrX_+_41433169
|
0.231
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr14_-_19867372
|
0.230
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr2_-_178495768
|
0.228
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr7_-_92615593
|
0.227
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr19_+_12064077
|
0.227
|
|
ZNF788
|
zinc finger family member 788
|
|
chr19_-_4509471
|
0.226
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr16_+_87687734
|
0.225
|
NM_001127214
NM_174917
|
ACSF3
|
acyl-CoA synthetase family member 3
|
|
chr1_+_51340487
|
0.225
|
NM_001136508
|
C1orf185
|
chromosome 1 open reading frame 185
|
|
chr1_+_235025162
|
0.223
|
NM_000254
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr13_+_48923688
|
0.223
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr11_+_59920062
|
0.223
|
NM_001079692
NM_032597
|
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14
|
|
chr14_-_20098929
|
0.223
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr10_-_21226536
|
0.223
|
NM_006393
|
NEBL
|
nebulette
|
|
chr3_-_127810087
|
0.222
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr6_+_132000134
|
0.220
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr19_+_21056792
|
0.220
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr2_-_190635566
|
0.220
|
NM_005259
|
MSTN
|
myostatin
|
|
chr6_+_26195426
|
0.220
|
NM_000410
NM_139003
NM_139004
NM_139006
NM_139007
NM_139008
NM_139009
NM_139010
NM_139011
|
HFE
|
hemochromatosis
|
|
chr1_+_235025313
|
0.219
|
|
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr1_-_44593522
|
0.217
|
NM_024066
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr10_+_13668964
|
0.217
|
|
PRPF18
|
PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae)
|
|
chr2_+_48698440
|
0.215
|
NM_001193487
NM_006872
|
GTF2A1L
|
general transcription factor IIA, 1-like
|
|
chr14_-_105997732
|
0.212
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_+_91762227
|
0.212
|
|
|
|
|
chr1_+_171871283
|
0.211
|
NM_001195190
|
LOC730159
|
hypothetical protein LOC730159
|
|
chr8_+_7821268
|
0.210
|
NM_001193630
|
LOC100132396
|
zinc finger protein 705D-like
|
|
chr19_+_15699833
|
0.208
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr1_-_39119845
|
0.208
|
NM_030772
|
GJA9-MYCBP
GJA9
|
GJA9-MYCBP read-through transcript
gap junction protein, alpha 9, 59kDa
|
|
chrX_+_47999740
|
0.207
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr4_-_38534802
|
0.207
|
NM_006068
|
TLR6
|
toll-like receptor 6
|