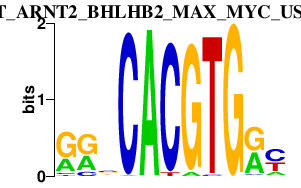
|
chr11_-_61441536
|
4.369
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr6_-_44333002
|
2.718
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr1_+_20385164
|
2.607
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr22_-_35232979
|
2.597
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr11_-_71492019
|
2.581
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_-_152797820
|
2.502
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr19_-_5291700
|
2.423
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr22_-_35233032
|
2.404
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr16_+_68542310
|
2.338
|
NM_182619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr16_+_68542126
|
2.329
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr6_+_32090516
|
2.250
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr19_+_54149963
|
2.233
|
|
BAX
|
BCL2-associated X protein
|
|
chr19_+_54149941
|
2.228
|
|
BAX
|
BCL2-associated X protein
|
|
chr17_+_73638430
|
2.226
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr19_+_54149843
|
2.144
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr11_-_71491942
|
1.997
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr11_-_71491983
|
1.990
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr7_+_150390566
|
1.897
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr6_+_32057779
|
1.885
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr12_-_56432385
|
1.879
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr6_-_33493792
|
1.828
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr6_-_33493898
|
1.819
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_-_27113018
|
1.754
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr6_+_87921980
|
1.654
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr1_-_152797767
|
1.630
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr17_+_37941476
|
1.617
|
NM_000263
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr6_-_43305156
|
1.601
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr12_-_56432292
|
1.594
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr6_-_31871394
|
1.587
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr9_-_137731139
|
1.584
|
NM_001012415
NM_001101677
|
SOHLH1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1
|
|
chr6_-_33494042
|
1.537
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_+_44870223
|
1.529
|
|
RNF220
|
ring finger protein 220
|
|
chr1_-_152797760
|
1.507
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr21_+_37367432
|
1.492
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr12_-_56432375
|
1.490
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr15_+_41871768
|
1.486
|
NM_001018108
|
SERF2
|
small EDRK-rich factor 2
|
|
chr11_-_71491916
|
1.485
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr1_+_109221271
|
1.479
|
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr3_+_33293915
|
1.472
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr12_-_56432377
|
1.424
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_+_14405216
|
1.416
|
|
PKN1
|
protein kinase N1
|
|
chr5_+_132237253
|
1.396
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr13_+_112999590
|
1.393
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr1_-_205186330
|
1.382
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr1_-_113300351
|
1.377
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr18_+_9698299
|
1.369
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr7_-_150847698
|
1.353
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr5_+_43156741
|
1.353
|
|
ZNF131
|
zinc finger protein 131
|
|
chr1_-_1700088
|
1.345
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr22_-_19543012
|
1.337
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr1_-_113300138
|
1.334
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr16_+_30951915
|
1.329
|
|
STX4
|
syntaxin 4
|
|
chr1_+_234916376
|
1.326
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr22_-_19543099
|
1.324
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr12_-_107551783
|
1.311
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr6_+_31976754
|
1.304
|
NM_001178063
|
C2
|
complement component 2
|
|
chr19_-_17236522
|
1.303
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr2_+_26840626
|
1.293
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr1_+_22650930
|
1.291
|
NM_001083621
NM_014870
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr10_-_12125028
|
1.291
|
NM_080599
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr22_-_19180075
|
1.286
|
NM_032775
|
KLHL22
|
kelch-like 22 (Drosophila)
|
|
chr7_-_150847529
|
1.284
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chrX_-_100490329
|
1.277
|
NM_001145951
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr17_-_44042924
|
1.255
|
|
HOXB7
|
homeobox B7
|
|
chr7_+_882704
|
1.253
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr17_+_37941790
|
1.248
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr1_+_1157485
|
1.244
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chr3_-_4483928
|
1.238
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chrX_-_100490612
|
1.231
|
NM_004085
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chr1_-_152797707
|
1.210
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr2_+_46779553
|
1.198
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr12_-_107551744
|
1.193
|
|
SELPLG
|
selectin P ligand
|
|
chrX_-_100549444
|
1.175
|
|
GLA
|
galactosidase, alpha
|
|
chr2_+_113119985
|
1.173
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr12_-_107551694
|
1.172
|
|
SELPLG
|
selectin P ligand
|
|
chr1_-_21868380
|
1.168
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr7_+_101860958
|
1.165
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr1_-_32054164
|
1.153
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr3_+_23962518
|
1.152
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_4483952
|
1.151
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr7_-_150847624
|
1.150
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr1_-_26266571
|
1.147
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr17_+_37941715
|
1.144
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr1_-_52936573
|
1.136
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr20_-_39199936
|
1.131
|
|
|
|
|
chr5_+_148766632
|
1.106
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr10_+_69314347
|
1.097
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chrX_-_100549561
|
1.093
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr18_+_148480
|
1.089
|
NM_001037334
NM_005151
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr7_+_101861007
|
1.083
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr2_+_177785667
|
1.083
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr19_+_37902518
|
1.078
|
NM_001110822
|
TDRD12
|
tudor domain containing 12
|
|
chr19_+_14405100
|
1.074
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr20_+_33823318
|
1.073
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr6_+_160310118
|
1.072
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr17_+_52518098
|
1.071
|
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr16_-_1465021
|
1.070
|
|
CLCN7
|
chloride channel 7
|
|
chr17_+_43374021
|
1.062
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr22_-_17798998
|
1.062
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_-_224195495
|
1.060
|
NM_001172425
NM_003240
|
LEFTY2
|
left-right determination factor 2
|
|
chr5_-_133734616
|
1.056
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr9_-_126309510
|
1.055
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr6_+_138230045
|
1.049
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr1_+_26609819
|
1.039
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr8_+_98857190
|
1.033
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr3_-_4483955
|
1.031
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr16_-_87244896
|
1.022
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr17_+_77528782
|
1.019
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr1_-_6585268
|
1.018
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr6_-_31871452
|
1.010
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr6_-_35764655
|
1.008
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr18_+_9698269
|
0.996
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr11_-_18300246
|
0.993
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr1_-_224178606
|
0.988
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr12_+_70434927
|
0.987
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr2_-_25328452
|
0.984
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr18_+_9698322
|
0.981
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr1_+_29086206
|
0.981
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_-_52604405
|
0.978
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr1_+_29086156
|
0.975
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_+_11718798
|
0.973
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr6_-_35764588
|
0.970
|
|
FKBP5
|
FK506 binding protein 5
|
|
chr2_-_172675873
|
0.968
|
|
DLX2
|
distal-less homeobox 2
|
|
chr1_+_22650934
|
0.964
|
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr14_+_99555471
|
0.961
|
|
EVL
|
Enah/Vasp-like
|
|
chr18_+_9698187
|
0.960
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr12_+_88627601
|
0.946
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr1_+_9522107
|
0.933
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr21_+_36458708
|
0.933
|
NM_005128
|
DOPEY2
|
dopey family member 2
|
|
chr1_-_52936602
|
0.922
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr1_+_117098579
|
0.918
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr15_+_41871809
|
0.918
|
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 read-through transcript
small EDRK-rich factor 2
|
|
chr6_-_28999388
|
0.904
|
|
TRIM27
|
tripartite motif containing 27
|
|
chr17_-_73636362
|
0.900
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr8_+_142471290
|
0.899
|
|
|
|
|
chr20_-_36191623
|
0.894
|
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr17_+_38186141
|
0.893
|
NM_032387
|
WNK4
|
WNK lysine deficient protein kinase 4
|
|
chr17_+_69781980
|
0.882
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr1_+_109963950
|
0.880
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_+_224083047
|
0.879
|
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr21_-_42247007
|
0.879
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr17_+_63713171
|
0.874
|
|
|
|
|
chr1_+_26368969
|
0.873
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr1_+_44870256
|
0.873
|
|
RNF220
|
ring finger protein 220
|
|
chr19_-_12693923
|
0.872
|
NM_013433
|
TNPO2
|
transportin 2
|
|
chr19_-_11549508
|
0.872
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr7_+_101704145
|
0.872
|
|
CUX1
|
cut-like homeobox 1
|
|
chr10_-_50066386
|
0.866
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr16_+_29726962
|
0.864
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr17_-_37586716
|
0.857
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr5_-_176869943
|
0.856
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr1_+_11718764
|
0.852
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr16_+_69171298
|
0.847
|
NM_001172771
NM_152456
|
IL34
|
interleukin 34
|
|
chr10_-_76665691
|
0.847
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr19_-_11549484
|
0.846
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr7_-_99612757
|
0.844
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr12_-_115660017
|
0.841
|
NM_024738
|
C12orf49
|
chromosome 12 open reading frame 49
|
|
chr10_+_64563009
|
0.841
|
NM_030759
|
NRBF2
|
nuclear receptor binding factor 2
|
|
chr17_-_35510074
|
0.837
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr6_-_31870809
|
0.834
|
|
|
|
|
chr10_+_45542653
|
0.830
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chrX_-_119593646
|
0.822
|
NM_003588
|
CUL4B
|
cullin 4B
|
|
chr18_+_75967902
|
0.822
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr1_-_180622083
|
0.821
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr9_+_114953048
|
0.820
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr19_+_10626106
|
0.817
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr15_-_99609197
|
0.811
|
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr1_-_40335496
|
0.811
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr22_-_19543069
|
0.810
|
NM_058004
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr19_-_47498768
|
0.809
|
NM_001145939
NM_001145940
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa)
|
|
chrX_+_49075036
|
0.808
|
NM_012196
NM_001127200
NM_001098412
|
GAGE8
GAGE2C
GAGE2E
GAGE13
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 13
G antigen 2A
|
|
chr20_+_43953372
|
0.807
|
|
CTSA
|
cathepsin A
|
|
chr16_+_24458349
|
0.806
|
NM_006910
NM_018703
NM_032626
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr7_+_106472243
|
0.804
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr7_-_150305933
|
0.790
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr3_+_4996305
|
0.789
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr2_+_119697853
|
0.787
|
NM_001008410
NM_018234
NM_182915
|
STEAP3
|
STEAP family member 3
|
|
chr3_+_184836049
|
0.787
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr1_+_205344200
|
0.786
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr1_+_11718715
|
0.785
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr10_-_45410197
|
0.777
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr9_+_114953087
|
0.776
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr11_+_18300370
|
0.774
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr20_+_43028533
|
0.774
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chrX_+_49241061
|
0.774
|
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 2A
|
|
chr1_-_40335504
|
0.774
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chrX_+_100549869
|
0.773
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_-_40335451
|
0.768
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr1_-_40335510
|
0.767
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr11_+_107384819
|
0.767
|
|
CUL5
|
cullin 5
|
|
chr19_+_10625987
|
0.767
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr4_+_609362
|
0.767
|
NM_000283
NM_001145291
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta
|
|
chr7_-_158304062
|
0.763
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr1_-_53380709
|
0.763
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chr17_+_32380287
|
0.762
|
NM_012138
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr1_-_28432095
|
0.762
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr19_-_10287673
|
0.761
|
NM_001031734
|
FDX1L
|
ferredoxin 1-like
|
|
chr20_-_60484420
|
0.759
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr9_-_139128983
|
0.755
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chrX_+_146870540
|
0.755
|
NM_152578
|
FMR1NB
|
fragile X mental retardation 1 neighbor
|