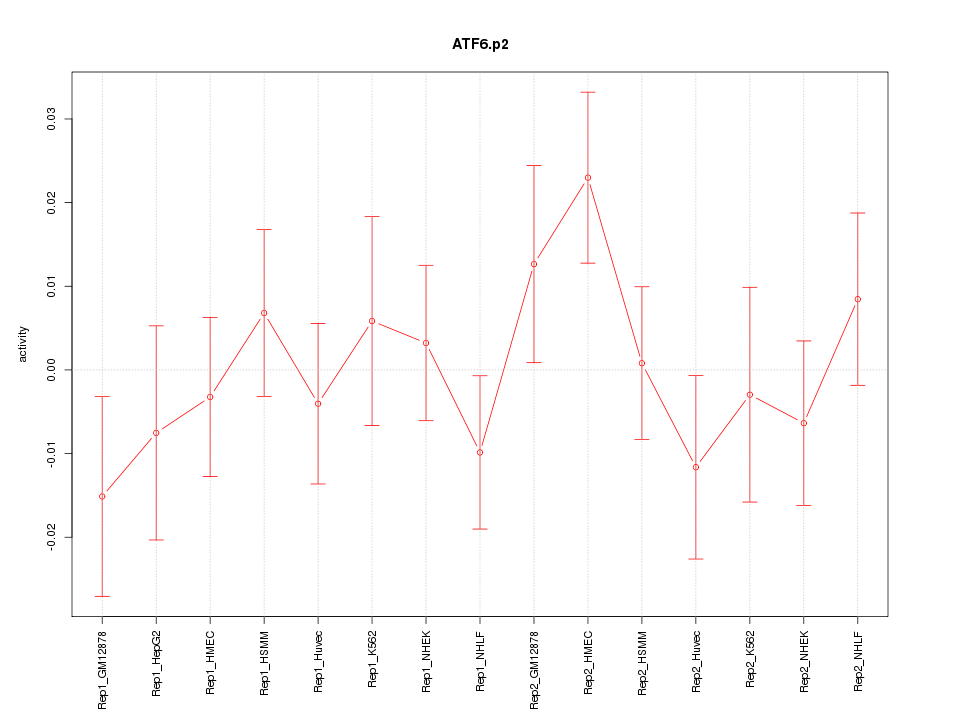
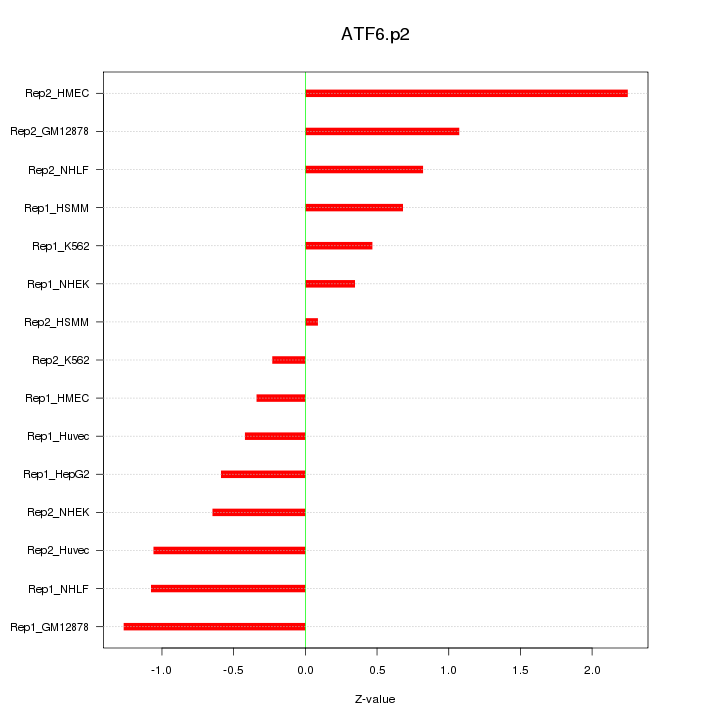
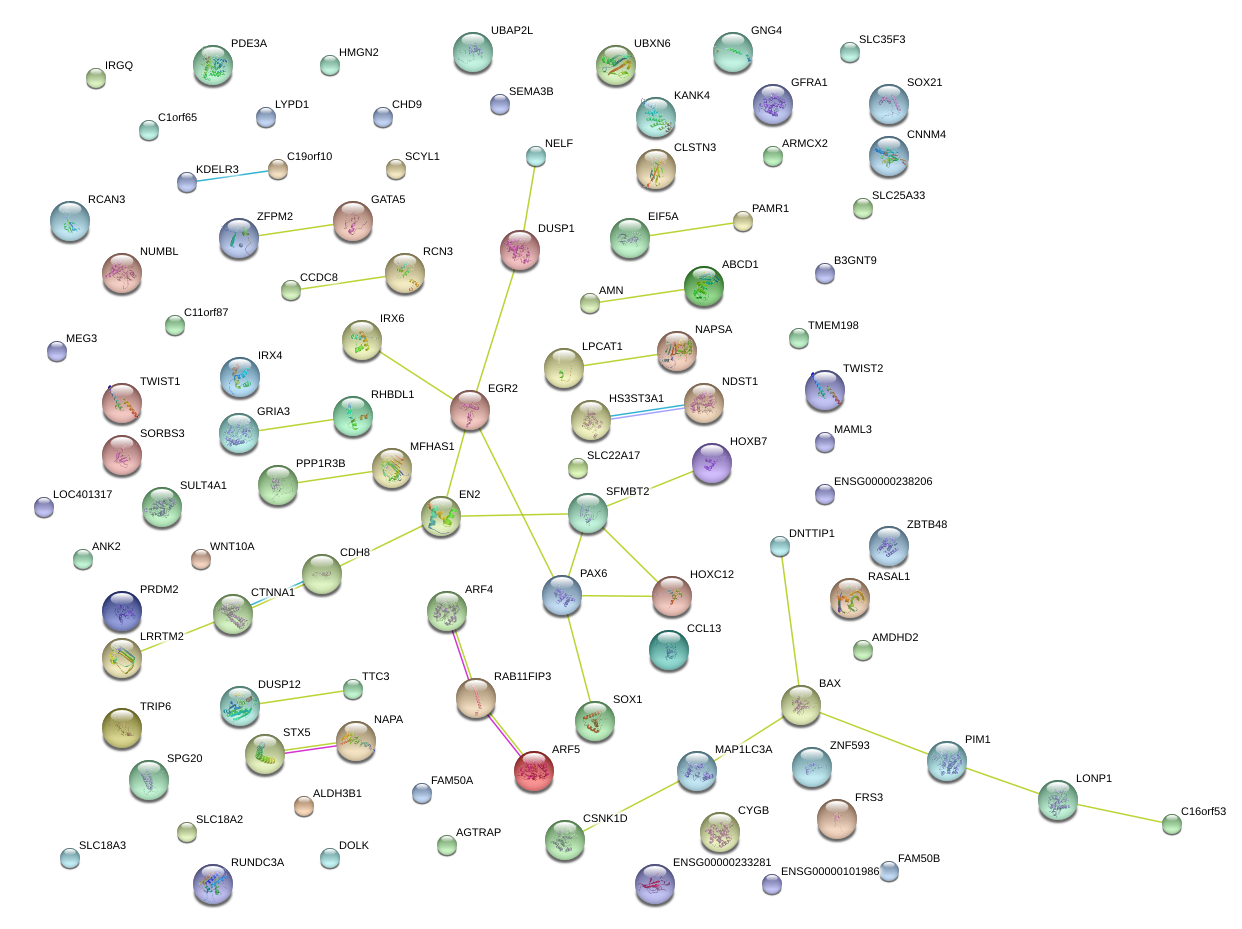
|
chr2_+_219453487
|
0.856
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr1_+_26368969
|
0.758
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr22_-_42589543
|
0.682
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr16_+_2510323
|
0.601
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr1_+_6562681
|
0.596
|
|
ZBTB48
|
zinc finger and BTB domain containing 48
|
|
chr17_-_44043278
|
0.554
|
|
HOXB7
|
homeobox B7
|
|
chr17_+_39741306
|
0.553
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr10_-_7491204
|
0.553
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr17_-_44043361
|
0.530
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr3_+_50281582
|
0.510
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr10_+_50488352
|
0.487
|
NM_003055
|
SLC18A3
|
solute carrier family 18 (vesicular acetylcholine), member 3
|
|
chr12_+_52634950
|
0.484
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr1_+_6562634
|
0.478
|
NM_005341
|
ZBTB48
|
zinc finger and BTB domain containing 48
|
|
chrX_+_122145776
|
0.455
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr5_-_172130766
|
0.414
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr5_+_78568094
|
0.411
|
|
|
|
|
chr1_-_233879617
|
0.409
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr11_+_65049552
|
0.400
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr16_+_415606
|
0.398
|
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr1_-_181654027
|
0.394
|
|
|
|
|
chr5_-_138238953
|
0.393
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr17_-_77824849
|
0.392
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr20_+_32610161
|
0.384
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr19_-_4621323
|
0.369
|
|
C19orf10
|
chromosome 19 open reading frame 10
|
|
chr20_-_60484420
|
0.367
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr8_-_11742668
|
0.366
|
|
|
|
|
chr17_-_72045218
|
0.364
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr8_+_145718702
|
0.361
|
|
|
|
|
chr9_-_130749604
|
0.359
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr5_-_138238891
|
0.357
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr19_+_54149963
|
0.357
|
|
BAX
|
BCL2-associated X protein
|
|
chr19_-_45888259
|
0.352
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr3_+_50281309
|
0.348
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr6_+_37245860
|
0.347
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chrX_-_100801472
|
0.344
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr16_-_65742323
|
0.342
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr5_-_1935764
|
0.341
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr16_+_29734990
|
0.335
|
NM_024516
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr16_-_60627536
|
0.334
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr12_+_110935612
|
0.332
|
|
|
|
|
chr7_+_154943466
|
0.331
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr17_-_13445942
|
0.331
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr11_-_31796061
|
0.330
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr2_-_133144094
|
0.327
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr4_-_141293572
|
0.326
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr8_-_9046460
|
0.324
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr11_-_35503720
|
0.318
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr16_+_51722285
|
0.313
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr5_+_149845503
|
0.312
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr9_-_139473606
|
0.312
|
NM_001130969
NM_001130970
NM_001130971
NM_001178064
NM_015537
|
NELF
|
nasal embryonic LHRH factor
|
|
chr5_-_1576908
|
0.311
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr19_+_54149941
|
0.309
|
|
BAX
|
BCL2-associated X protein
|
|
chr6_+_1336077
|
0.309
|
|
|
|
|
chr22_+_37193961
|
0.308
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr12_-_112058403
|
0.302
|
NM_001193520
NM_004658
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr14_+_102458745
|
0.299
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr2_+_220117009
|
0.298
|
|
TMEM198
|
transmembrane protein 198
|
|
chr1_+_152459918
|
0.297
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chrX_+_153325643
|
0.295
|
NM_004699
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr14_-_22891865
|
0.292
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr11_+_108798047
|
0.284
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr1_+_9522107
|
0.284
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr14_+_100362204
|
0.282
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr8_-_8788149
|
0.280
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr7_+_100302885
|
0.275
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr11_-_62356023
|
0.274
|
|
STX5
|
syntaxin 5
|
|
chr13_-_35818777
|
0.273
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr1_-_62557655
|
0.270
|
NM_181712
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr1_+_232416618
|
0.268
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr2_+_220116986
|
0.268
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr19_+_54723044
|
0.268
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr19_-_48791967
|
0.267
|
NM_001007561
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr8_+_106400322
|
0.267
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr19_-_4408789
|
0.266
|
NM_025241
|
UBXN6
|
UBX domain protein 6
|
|
chrX_+_153325684
|
0.265
|
|
FAM50A
|
family with sequence similarity 50, member A
|
|
chr1_+_13899272
|
0.263
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr12_+_7174230
|
0.262
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr9_-_139473433
|
0.260
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr4_+_114190233
|
0.259
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr7_+_28415495
|
0.259
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr13_-_94162249
|
0.258
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr11_+_67532592
|
0.254
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr19_+_54149843
|
0.251
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr15_-_61680003
|
0.251
|
|
LOC100130855
|
hypothetical LOC100130855
|
|
chr17_+_7151653
|
0.249
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr3_-_57558120
|
0.248
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr17_+_77478858
|
0.248
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr16_+_53915971
|
0.247
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr11_-_67329246
|
0.246
|
|
LOC645332
|
family with sequence similarity 86, member A pseudogene
|
|
chr20_+_43854013
|
0.246
|
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr14_+_100362236
|
0.246
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr10_-_64246129
|
0.246
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr19_-_52709975
|
0.245
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_+_26671488
|
0.244
|
NM_005517
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr8_+_22479115
|
0.242
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr1_+_11718715
|
0.242
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr17_+_7152417
|
0.241
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr1_+_9522133
|
0.241
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr10_+_118990573
|
0.240
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr21_+_37367432
|
0.239
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr20_+_43853982
|
0.239
|
NM_052951
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr7_-_19123765
|
0.239
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr2_+_96790298
|
0.239
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr10_-_118022965
|
0.239
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr9_-_139473569
|
0.238
|
|
NELF
|
nasal embryonic LHRH factor
|
|
chr12_+_20413445
|
0.237
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr3_-_57558130
|
0.237
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr16_+_665680
|
0.237
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr19_-_51608665
|
0.236
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr1_+_221633337
|
0.236
|
NM_152610
|
C1orf65
|
chromosome 1 open reading frame 65
|
|
chr1_-_114497994
|
0.235
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr12_-_14612057
|
0.234
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr17_+_77528782
|
0.233
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr16_+_29726931
|
0.232
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr21_-_43671355
|
0.231
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr2_-_192767844
|
0.230
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr11_-_59139755
|
0.230
|
|
OSBP
|
oxysterol binding protein
|
|
chr4_+_1232942
|
0.230
|
|
C4orf42
|
chromosome 4 open reading frame 42
|
|
chr17_-_77478835
|
0.227
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr1_+_11718764
|
0.226
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr1_+_220857774
|
0.226
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr5_-_1577022
|
0.225
|
NM_024830
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr11_-_65138217
|
0.224
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr11_-_118740069
|
0.224
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr12_+_52665196
|
0.224
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr12_+_70120028
|
0.223
|
NM_003667
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr7_+_5598964
|
0.222
|
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr16_+_29734785
|
0.222
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr11_+_7491789
|
0.221
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr2_-_96899396
|
0.221
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr19_-_17660005
|
0.220
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr12_+_2856622
|
0.218
|
|
C12orf32
|
chromosome 12 open reading frame 32
|
|
chr1_-_181654124
|
0.217
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr19_-_62912347
|
0.216
|
NM_001085384
|
ZNF154
|
zinc finger protein 154
|
|
chr12_-_112394259
|
0.215
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr12_+_54838408
|
0.214
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr9_-_78710698
|
0.214
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr12_+_110935534
|
0.214
|
NM_001034025
NM_006817
|
ERP29
|
endoplasmic reticulum protein 29
|
|
chr15_-_80125403
|
0.213
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr2_+_28469199
|
0.212
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr1_-_226357552
|
0.212
|
NM_024319
|
C1orf35
|
chromosome 1 open reading frame 35
|
|
chr20_-_4177519
|
0.211
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr1_-_201194033
|
0.211
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr7_+_100109359
|
0.211
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr14_+_103065288
|
0.211
|
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr17_-_34157962
|
0.209
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr11_-_118739838
|
0.209
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_62557559
|
0.208
|
|
KANK4
|
KN motif and ankyrin repeat domains 4
|
|
chr17_-_53439570
|
0.208
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr14_-_102045749
|
0.207
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr12_+_6768960
|
0.207
|
|
CD4
|
CD4 molecule
|
|
chr1_+_159390379
|
0.207
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr17_-_53439589
|
0.207
|
|
SRSF1
|
serine/arginine-rich splicing factor 1
|
|
chr12_-_14611820
|
0.205
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr2_+_28469172
|
0.204
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_150414704
|
0.201
|
NM_001042535
NM_031946
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr17_-_32368033
|
0.201
|
|
|
|
|
chr15_-_77169894
|
0.200
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr19_+_5632016
|
0.199
|
NM_198704
NM_198705
NM_198533
NM_198707
NM_198706
NM_198708
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr10_-_88116181
|
0.199
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr11_-_65138323
|
0.199
|
|
|
|
|
chr11_+_108798084
|
0.199
|
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr19_-_52710089
|
0.199
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr1_+_110494654
|
0.198
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr17_+_77529070
|
0.198
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr7_-_19123787
|
0.198
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr3_+_45959976
|
0.198
|
NM_006564
|
CXCR6
|
chemokine (C-X-C motif) receptor 6
|
|
chr1_+_109810743
|
0.198
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr15_-_66511488
|
0.198
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr5_-_34020445
|
0.196
|
NM_001012509
NM_016180
|
SLC45A2
|
solute carrier family 45, member 2
|
|
chr1_-_172103745
|
0.196
|
|
GAS5
|
growth arrest-specific 5 (non-protein coding)
|
|
chr14_-_22458199
|
0.194
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr22_-_19180075
|
0.193
|
NM_032775
|
KLHL22
|
kelch-like 22 (Drosophila)
|
|
chr2_-_27384617
|
0.193
|
NM_003353
|
UCN
|
urocortin
|
|
chr22_+_20101661
|
0.193
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr11_-_62356094
|
0.193
|
NM_003164
|
STX5
|
syntaxin 5
|
|
chrX_-_139414890
|
0.193
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr15_-_80125514
|
0.192
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr9_+_86474500
|
0.192
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr12_+_6768934
|
0.192
|
NM_001195014
NM_001195015
NM_001195016
NM_001195017
|
CD4
|
CD4 molecule
|
|
chr7_+_100109245
|
0.191
|
NM_005273
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr1_-_50661716
|
0.191
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr1_+_109810622
|
0.190
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr9_-_103289154
|
0.190
|
NM_032342
|
C9orf125
|
chromosome 9 open reading frame 125
|
|
chr10_-_128067062
|
0.190
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_-_201194083
|
0.189
|
NM_015999
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr8_+_142471290
|
0.188
|
|
|
|
|
chr20_-_43853945
|
0.188
|
NM_080614
|
WFDC3
|
WAP four-disulfide core domain 3
|
|
chr12_+_11694172
|
0.187
|
|
ETV6
|
ets variant 6
|
|
chrX_-_55532355
|
0.187
|
NM_201286
|
USP51
|
ubiquitin specific peptidase 51
|
|
chr20_-_2769245
|
0.186
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr12_+_6768966
|
0.185
|
|
CD4
|
CD4 molecule
|
|
chr5_-_176760242
|
0.185
|
NM_001029886
|
PFN3
|
profilin 3
|
|
chr10_+_119291945
|
0.185
|
NM_001165924
NM_004098
|
EMX2
|
empty spiracles homeobox 2
|
|
chr8_+_142207866
|
0.185
|
NM_014957
|
DENND3
|
DENN/MADD domain containing 3
|
|
chr19_-_52710129
|
0.185
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chrX_+_101267315
|
0.184
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chrX_+_133952633
|
0.184
|
NM_001163438
|
LOC644538
|
hypothetical protein LOC644538
|
|
chr1_+_226337016
|
0.184
|
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr1_-_43509187
|
0.183
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|