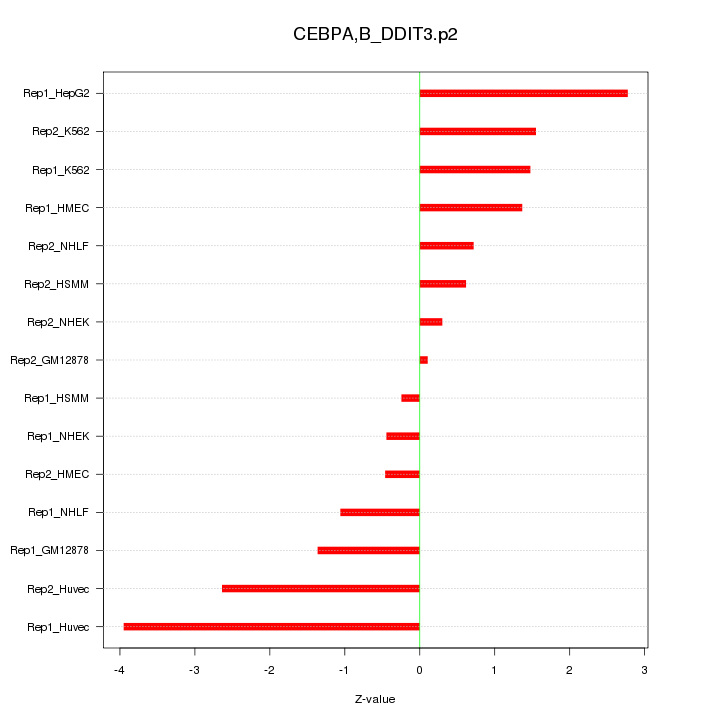
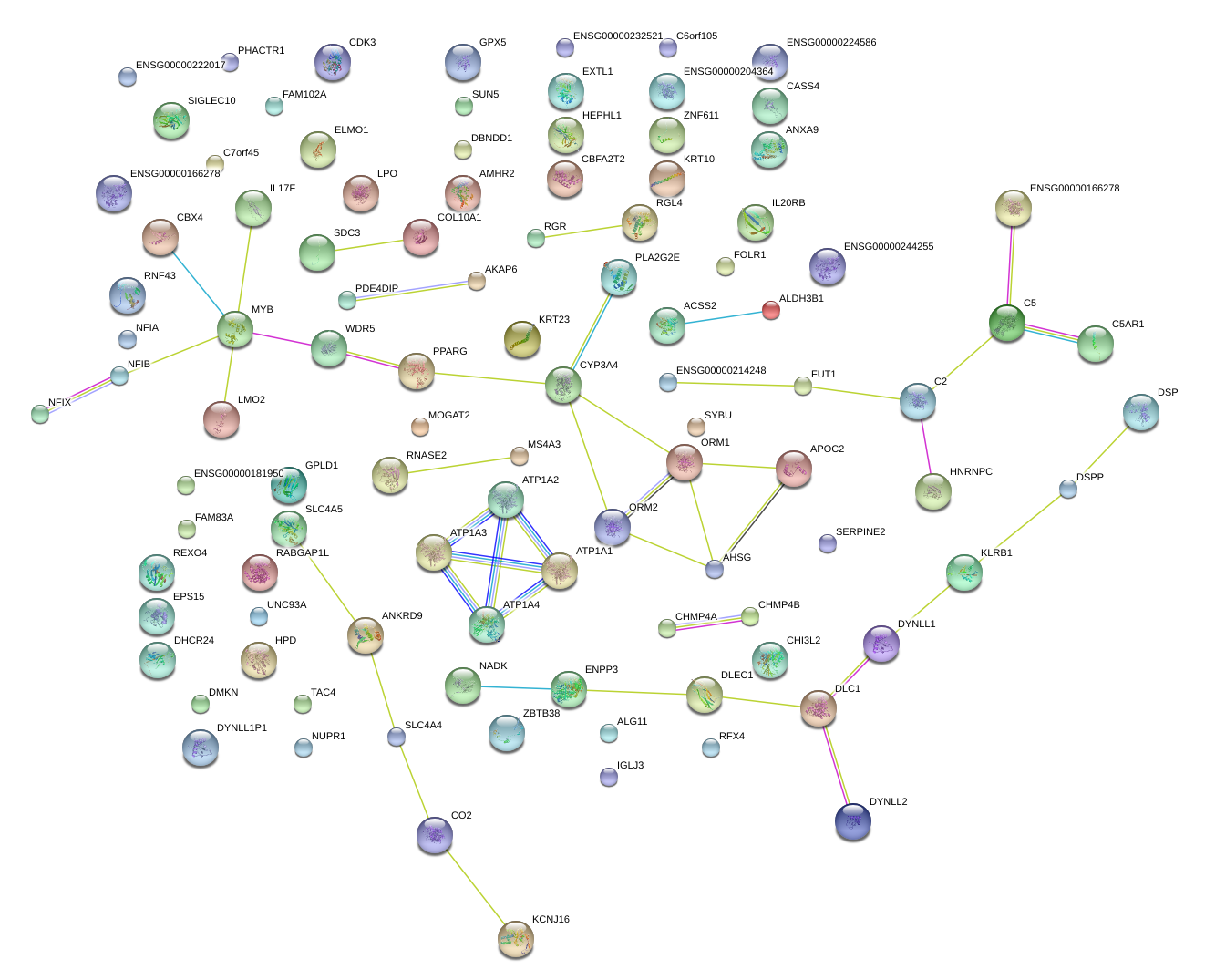
|
chr1_+_149221122
|
8.324
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr19_-_53950458
|
3.569
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr1_+_173111306
|
2.780
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr1_-_20122638
|
2.715
|
NM_014589
|
PLA2G2E
|
phospholipase A2, group IIE
|
|
chr17_-_36346240
|
2.647
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_+_167624792
|
2.453
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr9_+_116125123
|
2.381
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr17_+_71508581
|
2.294
|
NM_001258
|
CDK3
|
cyclin-dependent kinase 3
|
|
chr6_+_32003380
|
2.211
|
|
C2
|
complement component 2
|
|
chr1_-_143708467
|
2.118
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_1701313
|
2.117
|
NM_001198994
|
NADK
|
NAD kinase
|
|
chr3_-_42818221
|
2.047
|
|
|
|
|
chr3_+_142525744
|
1.868
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr6_+_32003180
|
1.851
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr1_+_111571803
|
1.800
|
NM_001025197
NM_004000
|
CHI3L2
|
chitinase 3-like 2
|
|
chr22_+_21393107
|
1.713
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr10_+_85994788
|
1.708
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr3_+_187813538
|
1.683
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr8_+_124263932
|
1.596
|
NM_032899
NM_207006
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr6_-_24597717
|
1.588
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr1_+_26220895
|
1.579
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr13_+_51496679
|
1.562
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr3_+_187813405
|
1.554
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr11_+_67534364
|
1.538
|
NM_000694
NM_001030010
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr14_+_31868229
|
1.504
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr17_-_45280377
|
1.478
|
NM_001077503
NM_001077504
NM_001077505
NM_001077506
NM_170685
|
TAC4
|
tachykinin 4 (hemokinin)
|
|
chr16_-_28457728
|
1.421
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr12_+_105519049
|
1.419
|
NM_002920
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr1_-_31117948
|
1.395
|
|
SDC3
|
syndecan 3
|
|
chr11_+_93394025
|
1.392
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr3_+_138159396
|
1.372
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr11_+_67534371
|
1.370
|
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr1_+_116744992
|
1.303
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr8_+_124264054
|
1.296
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr14_+_20493467
|
1.263
|
NM_002934
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin)
|
|
chr9_-_70780574
|
1.252
|
|
|
|
|
chr16_-_28457825
|
1.251
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr19_+_46121963
|
1.244
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr6_+_37511383
|
1.223
|
|
|
|
|
chr9_-_129752687
|
1.220
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr12_+_52105131
|
1.204
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr17_-_36232366
|
1.191
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr11_+_59580630
|
1.181
|
NM_001031666
NM_001031809
NM_006138
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr1_-_55103938
|
1.150
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr19_+_50144267
|
1.145
|
|
APOC2
|
apolipoprotein C-II
|
|
chr11_+_59580705
|
1.142
|
|
MS4A3
|
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific)
|
|
chr6_-_52217185
|
1.128
|
NM_052872
|
IL17F
|
interleukin 17F
|
|
chr11_-_33870411
|
1.121
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr19_-_40684638
|
1.108
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr9_-_14076901
|
1.086
|
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_28601767
|
1.084
|
NM_001509
NM_003996
|
GPX5
|
glutathione peroxidase 5 (epididymal androgen-related protein)
|
|
chr3_+_187813553
|
1.069
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr17_+_53670785
|
1.061
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr1_-_51660328
|
1.061
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr19_-_57924946
|
1.051
|
NM_030972
|
ZNF611
|
zinc finger protein 611
|
|
chr11_+_75106512
|
1.036
|
NM_025098
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chr3_+_12367993
|
1.032
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr6_-_11887265
|
1.030
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chrX_-_24241352
|
0.963
|
NM_001136233
|
FAM48B2
|
family with sequence similarity 48, member B2
|
|
chr14_-_23752875
|
0.941
|
NM_014169
|
CHMP4A
|
chromatin modifying protein 4A
|
|
chr12_-_120781002
|
0.930
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr6_+_12825022
|
0.901
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr20_+_54420574
|
0.888
|
NM_001164116
NM_001164114
NM_001164115
NM_020356
|
CASS4
|
Cas scaffolding protein family member 4
|
|
chr6_+_135558572
|
0.880
|
|
|
|
|
chr7_+_129634935
|
0.876
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr17_-_75427644
|
0.867
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr7_-_36991189
|
0.852
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_+_52504914
|
0.851
|
NM_001736
|
C5AR1
|
complement component 5a receptor 1
|
|
chr20_-_31055892
|
0.840
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr19_-_56612653
|
0.834
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr16_-_88604029
|
0.832
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr1_+_559129
|
0.800
|
|
|
|
|
chr8_-_13018123
|
0.798
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_+_12824873
|
0.792
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr10_-_15237351
|
0.789
|
|
LOC100192204
|
peptidylprolyl isomerase A pseudogene
|
|
chr9_-_122852356
|
0.767
|
NM_001735
|
C5
|
complement component 5
|
|
chr6_+_135558744
|
0.764
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_53847937
|
0.764
|
|
RNF43
|
ring finger protein 43
|
|
chr6_-_116553918
|
0.752
|
NM_000493
|
COL10A1
|
collagen, type X, alpha 1
|
|
chr2_-_74395657
|
0.743
|
NM_021196
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr2_-_224574860
|
0.731
|
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr9_+_135994734
|
0.725
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr20_+_31613800
|
0.717
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr1_+_158387975
|
0.717
|
NM_144699
|
ATP1A4
|
ATPase, Na+/K+ transporting, alpha 4 polypeptide
|
|
chr6_+_158654890
|
0.716
|
|
|
|
|
chr8_-_110730183
|
0.705
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr11_+_71580816
|
0.703
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr6_+_132000134
|
0.695
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr20_+_32926452
|
0.693
|
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr17_+_65582981
|
0.691
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr6_+_7524691
|
0.675
|
|
DSP
|
desmoplakin
|
|
chr12_-_53153639
|
0.669
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr12_+_54910086
|
0.661
|
NM_173596
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr3_-_49701085
|
0.661
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr7_-_92686773
|
0.658
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr2_+_169465995
|
0.650
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr12_+_27740694
|
0.650
|
NM_001029874
|
REP15
|
RAB15 effector protein
|
|
chr19_+_11211282
|
0.649
|
NM_018687
|
LOC55908
|
hepatocellular carcinoma-associated gene TD26
|
|
chr7_+_99263571
|
0.642
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr6_+_131936057
|
0.636
|
NM_000045
|
ARG1
|
arginase, liver
|
|
chr11_+_2279932
|
0.630
|
|
TSPAN32
|
tetraspanin 32
|
|
chr14_-_69333624
|
0.622
|
NM_003049
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr2_-_121964373
|
0.597
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr9_-_14769530
|
0.594
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr22_+_21079355
|
0.593
|
|
|
|
|
chr1_-_120155581
|
0.579
|
NM_001159352
NM_001159353
NM_032044
|
REG4
|
regenerating islet-derived family, member 4
|
|
chr2_-_21084911
|
0.577
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_+_159943385
|
0.558
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr11_+_47236072
|
0.556
|
NM_001130101
NM_005693
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chrX_-_152748526
|
0.554
|
|
PDZD4
|
PDZ domain containing 4
|
|
chr22_+_20880159
|
0.546
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr1_-_180822686
|
0.542
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr11_+_71524403
|
0.535
|
NM_000804
|
FOLR3
|
folate receptor 3 (gamma)
|
|
chr1_+_26376611
|
0.530
|
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr3_-_9570412
|
0.529
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr7_-_111216180
|
0.527
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr11_-_116168316
|
0.523
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr1_+_234748187
|
0.523
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr12_+_99391870
|
0.516
|
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr12_-_55931172
|
0.506
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr2_+_191009265
|
0.505
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr22_+_19248810
|
0.502
|
|
MED15
|
mediator complex subunit 15
|
|
chr22_+_24174039
|
0.501
|
|
CRYBB2P1
|
crystallin, beta B2 pseudogene 1
|
|
chr8_-_66863841
|
0.500
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr1_+_64442077
|
0.498
|
NM_152489
|
UBE2U
|
ubiquitin-conjugating enzyme E2U (putative)
|
|
chr8_-_17787342
|
0.496
|
|
FGL1
|
fibrinogen-like 1
|
|
chr16_+_3345889
|
0.495
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr17_+_30472710
|
0.493
|
NM_017559
|
FNDC8
|
fibronectin type III domain containing 8
|
|
chr4_-_26100986
|
0.489
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr11_+_62893836
|
0.489
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr3_-_150422474
|
0.486
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr1_-_172153024
|
0.484
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr11_-_10487212
|
0.481
|
NM_001190702
|
MTRNR2L8
|
MT-RNR2-like 8
|
|
chr7_-_112515035
|
0.475
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr14_+_73153300
|
0.470
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chr19_-_12858956
|
0.469
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr6_+_31604717
|
0.459
|
NM_001011700
|
MCCD1
|
mitochondrial coiled-coil domain 1
|
|
chrX_-_13745108
|
0.458
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_+_233605625
|
0.457
|
NM_005383
|
NEU2
|
sialidase 2 (cytosolic sialidase)
|
|
chr3_+_187917791
|
0.455
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr9_+_95886625
|
0.452
|
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chrX_-_70243362
|
0.448
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr17_-_15463742
|
0.448
|
NM_006382
|
CDRT1
|
CMT1A duplicated region transcript 1
|
|
chr6_+_26195426
|
0.447
|
NM_000410
NM_139003
NM_139004
NM_139006
NM_139007
NM_139008
NM_139009
NM_139010
NM_139011
|
HFE
|
hemochromatosis
|
|
chr1_+_557269
|
0.445
|
|
|
|
|
chr9_+_94766379
|
0.445
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr1_+_151596953
|
0.444
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chrX_+_44588192
|
0.442
|
NM_022076
|
DUSP21
|
dual specificity phosphatase 21
|
|
chr19_-_4253389
|
0.441
|
NM_001169126
NM_144615
|
TMIGD2
|
transmembrane and immunoglobulin domain containing 2
|
|
chr22_+_20880112
|
0.436
|
|
|
|
|
chr17_+_18588050
|
0.430
|
NM_031456
|
FBXW10
CDRT1
|
F-box and WD repeat domain containing 10
CMT1A duplicated region transcript 1
|
|
chr1_-_21373816
|
0.429
|
NM_001198803
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr11_+_73339011
|
0.422
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr17_-_77498399
|
0.416
|
NM_001145113
|
MYADML2
|
myeloid-associated differentiation marker-like 2
|
|
chr9_-_116151099
|
0.412
|
|
AKNA
|
AT-hook transcription factor
|
|
chr15_+_60838053
|
0.412
|
|
TLN2
|
talin 2
|
|
chr17_-_17426416
|
0.410
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_+_17779528
|
0.408
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr7_-_104354325
|
0.400
|
|
LOC723809
|
hypothetical LOC723809
|
|
chrX_+_77267477
|
0.396
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr9_+_109085386
|
0.396
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr20_+_309272
|
0.391
|
NM_021158
|
TRIB3
|
tribbles homolog 3 (Drosophila)
|
|
chr17_+_3326045
|
0.389
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr22_+_29848923
|
0.383
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr6_+_64414389
|
0.382
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr19_+_4442610
|
0.382
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr19_-_43926953
|
0.373
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr2_+_10103735
|
0.372
|
|
|
|
|
chr1_-_176205672
|
0.368
|
NM_033127
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr19_-_59368625
|
0.367
|
NM_001145303
NM_144686
|
TMC4
|
transmembrane channel-like 4
|
|
chr19_+_53770804
|
0.364
|
NM_004605
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chrX_-_15198509
|
0.363
|
NM_001168531
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr5_-_176769094
|
0.361
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr6_-_31954745
|
0.358
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr6_-_42270671
|
0.351
|
NM_002098
|
GUCA1B
|
guanylate cyclase activator 1B (retina)
|
|
chr8_-_86477585
|
0.347
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr15_+_56511466
|
0.347
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr17_+_1612056
|
0.347
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chrX_-_13745134
|
0.341
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_+_12775814
|
0.338
|
|
|
|
|
chr1_+_26376560
|
0.338
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr21_-_42689192
|
0.338
|
NM_024022
NM_032405
|
TMPRSS3
|
transmembrane protease, serine 3
|
|
chr15_+_83228895
|
0.334
|
NM_004213
NM_201651
|
SLC28A1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1
|
|
chr1_-_57057956
|
0.332
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chrX_+_69314060
|
0.329
|
NM_198512
|
DGAT2L6
|
diacylglycerol O-acyltransferase 2-like 6
|
|
chr10_+_70650064
|
0.328
|
NM_025130
|
HKDC1
|
hexokinase domain containing 1
|
|
chr8_+_26569709
|
0.328
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_53787187
|
0.328
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr6_-_49862858
|
0.325
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr8_-_27997306
|
0.324
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr17_+_1612031
|
0.323
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr11_-_614901
|
0.322
|
NM_001171968
NM_021924
NM_031264
|
CDHR5
|
cadherin-related family member 5
|
|
chr11_+_5558682
|
0.321
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr2_-_21081694
|
0.320
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr16_-_30301634
|
0.320
|
NM_052838
|
SEPT1
|
septin 1
|
|
chrX_+_106030049
|
0.319
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr1_-_44593522
|
0.318
|
NM_024066
|
ERI3
|
ERI1 exoribonuclease family member 3
|
|
chr19_+_48912053
|
0.317
|
NM_019612
|
IRGC
|
immunity-related GTPase family, cinema
|
|
chr3_+_188222337
|
0.315
|
NM_003032
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr11_+_62813947
|
0.315
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|