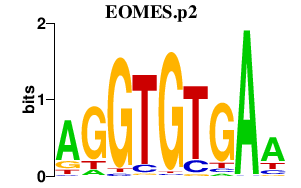
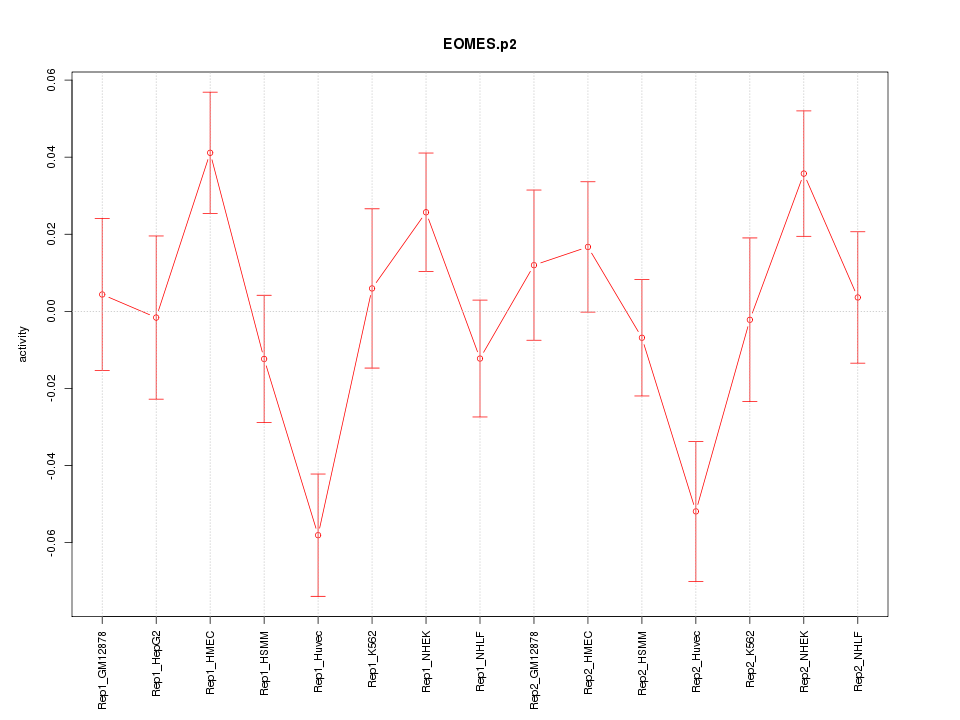
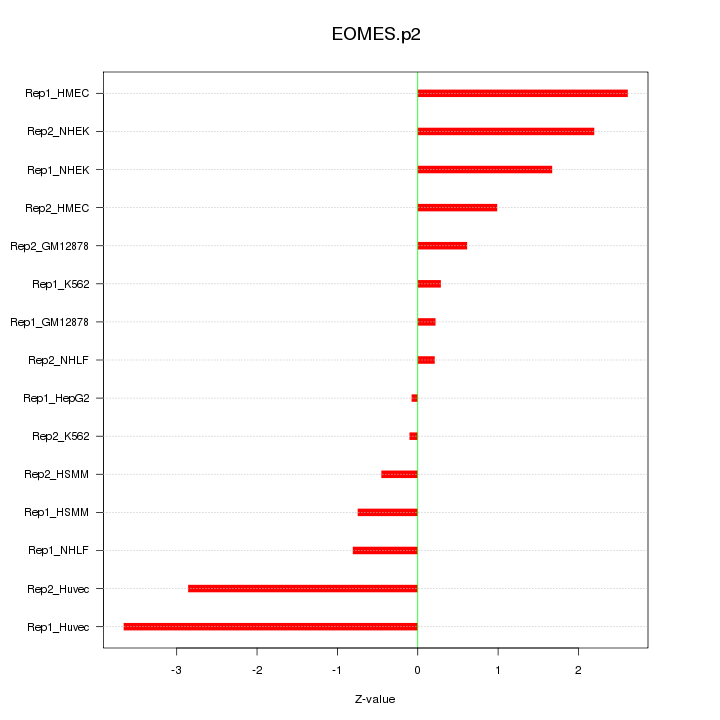
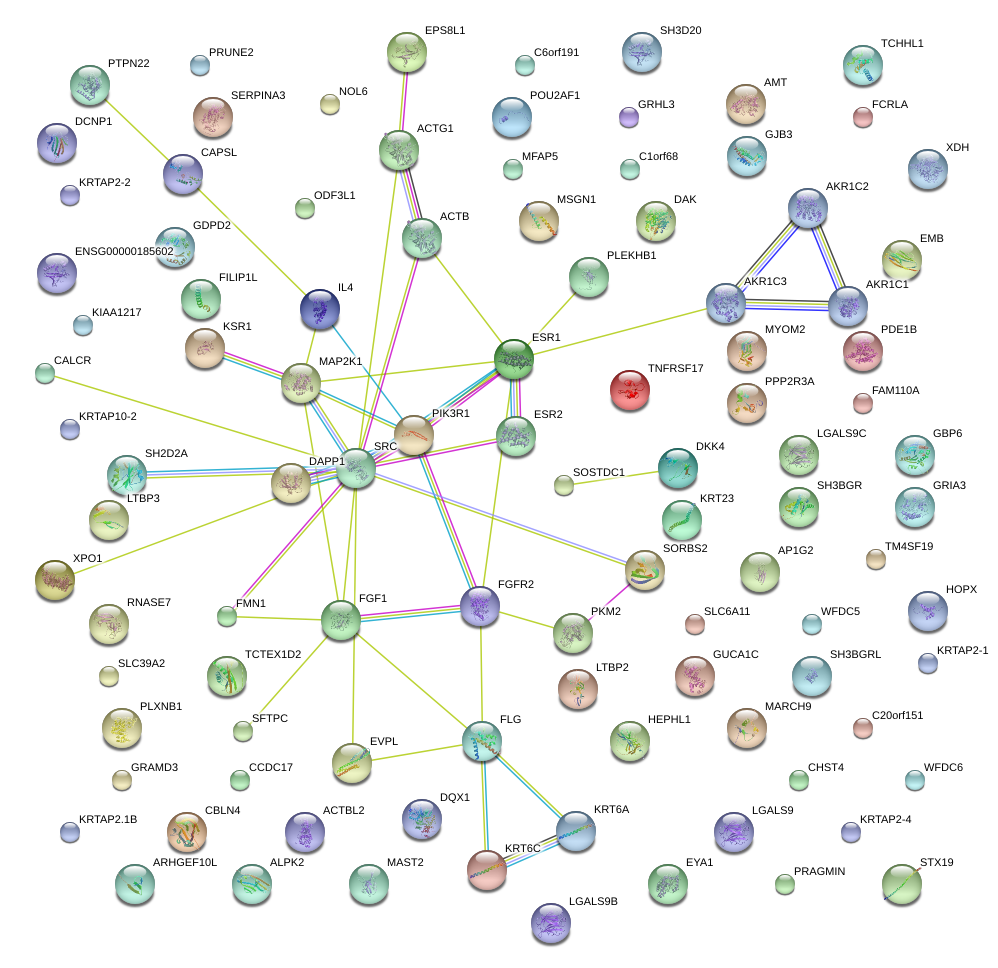
|
chr2_-_74606819
|
4.116
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr1_+_150958621
|
3.282
|
NM_001024679
|
C1orf68
|
chromosome 1 open reading frame 68
|
|
chr17_-_20311411
|
2.792
|
NM_001042685
|
LGALS9B
|
lectin, galactoside-binding, soluble, 9B
|
|
chr17_+_22823151
|
2.758
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr12_-_51173238
|
2.631
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr1_+_17719405
|
2.547
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr17_+_18320822
|
2.474
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr8_-_11742668
|
2.336
|
|
|
|
|
chr11_+_93394025
|
2.248
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr17_-_40858775
|
2.141
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr1_+_89602023
|
2.085
|
NM_198460
|
GBP6
|
guanylate binding protein family, member 6
|
|
chr8_-_72431274
|
1.997
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr3_+_137224266
|
1.939
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr11_-_65070625
|
1.937
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr17_-_36475652
|
1.917
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr3_-_48446405
|
1.898
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr1_+_24522116
|
1.890
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr3_-_49434874
|
1.874
|
|
AMT
|
aminomethyltransferase
|
|
chr14_+_20537253
|
1.786
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr9_-_33455191
|
1.767
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr10_-_123346148
|
1.751
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_-_36469869
|
1.744
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr3_-_110155237
|
1.700
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr15_-_70288071
|
1.593
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr8_+_22075112
|
1.542
|
NM_001172357
NM_001172410
NM_003018
|
SFTPC
|
surfactant protein C
|
|
chr6_-_130224108
|
1.521
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr3_+_10832867
|
1.492
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr5_-_56814310
|
1.464
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr7_-_16471817
|
1.423
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr6_+_152168500
|
1.422
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr2_+_17861266
|
1.372
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr1_+_35019376
|
1.352
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr20_+_35406497
|
1.349
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chrX_+_69559605
|
1.308
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr21_+_39745649
|
1.285
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr10_+_4995685
|
1.268
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr14_+_94148466
|
1.263
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr15_+_64481270
|
1.247
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr10_+_4995597
|
1.240
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr3_-_95230143
|
1.196
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr16_+_11966464
|
1.150
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr15_+_73803351
|
1.148
|
NM_175881
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr12_-_8706637
|
1.137
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chrX_+_122145776
|
1.102
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr5_+_132037271
|
1.079
|
NM_000589
NM_172348
|
IL4
|
interleukin 4
|
|
chrX_-_62564934
|
1.078
|
|
LOC92249
|
hypothetical LOC92249
|
|
chr18_-_54447168
|
1.061
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr20_+_773283
|
1.060
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr5_+_67558217
|
1.053
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr3_-_101316038
|
1.007
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr8_-_42353719
|
1.004
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr8_+_1980564
|
1.003
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chrX_-_135783496
|
1.001
|
|
|
|
|
chr2_-_31491057
|
0.994
|
|
XDH
|
xanthine dehydrogenase
|
|
chr20_-_43177137
|
0.993
|
NM_145652
|
WFDC5
|
WAP four-disulfide core domain 5
|
|
chr16_+_70117523
|
0.985
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr1_-_150328153
|
0.958
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr4_+_100957003
|
0.954
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr1_-_155053124
|
0.941
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr8_-_8276598
|
0.941
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr9_-_78710698
|
0.940
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr17_+_22982319
|
0.937
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr10_+_24538094
|
0.931
|
|
KIAA1217
|
KIAA1217
|
|
chr17_+_22982278
|
0.925
|
NM_002308
NM_009587
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr4_-_186969202
|
0.907
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_-_23106372
|
0.895
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr17_+_22982344
|
0.893
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr1_+_159943385
|
0.890
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr5_-_141973875
|
0.884
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr11_+_73036241
|
0.883
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr17_+_22982372
|
0.880
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr17_+_22982383
|
0.876
|
|
LGALS9
|
lectin, galactoside-binding, soluble, 9
|
|
chr12_+_56435042
|
0.864
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr11_+_60858611
|
0.851
|
|
DAK
|
dihydroxyacetone kinase 2 homolog (S. cerevisiae)
|
|
chr1_-_45862225
|
0.840
|
NM_001114938
NM_001190182
|
CCDC17
|
coiled-coil domain containing 17
|
|
chr15_-_31147376
|
0.836
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr5_-_49772973
|
0.835
|
NM_198449
|
EMB
|
embigin
|
|
chr17_-_36346240
|
0.834
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr20_-_60435983
|
0.826
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr12_+_53229625
|
0.826
|
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr21_-_44795769
|
0.825
|
NM_198693
|
KRTAP10-2
|
keratin associated protein 10-2
|
|
chr20_-_54013418
|
0.818
|
NM_080617
|
CBLN4
|
cerebellin 4 precursor
|
|
chr11_-_110755112
|
0.807
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr20_-_43601542
|
0.801
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr14_+_20580224
|
0.799
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr2_-_31491102
|
0.793
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr7_-_93027057
|
0.777
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr1_+_46151845
|
0.777
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr9_+_126460511
|
0.753
|
|
|
|
|
chr5_-_134810696
|
0.752
|
NM_130848
|
C5orf20
|
chromosome 5 open reading frame 20
|
|
chr1_-_43523825
|
0.751
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr5_-_35974493
|
0.750
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr3_-_197549588
|
0.749
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr17_-_71535076
|
0.748
|
NM_001988
|
EVPL
|
envoplakin
|
|
chr19_+_60279032
|
0.747
|
NM_133180
|
EPS8L1
|
EPS8-like 1
|
|
chr1_-_114215768
|
0.743
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr5_+_125828685
|
0.742
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr4_-_57242492
|
0.739
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr21_-_30774502
|
0.737
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr4_-_186969041
|
0.724
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr16_+_2757655
|
0.708
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_+_9132367
|
0.697
|
|
|
|
|
chr7_-_36730525
|
0.691
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr8_+_62363078
|
0.688
|
NM_173519
|
CLVS1
|
clavesin 1
|
|
chr6_-_31246159
|
0.684
|
NM_002701
|
POU5F1
POU5F1B
POU5F1P3
|
POU class 5 homeobox 1
POU class 5 homeobox 1B
POU class 5 homeobox 1 pseudogene 3
|
|
chr2_-_215711313
|
0.677
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr10_-_105228986
|
0.671
|
NM_001129742
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr4_+_75393053
|
0.655
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr12_+_53229443
|
0.655
|
NM_000924
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr6_+_64339869
|
0.654
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_-_159306037
|
0.654
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr11_-_62445587
|
0.651
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr11_-_46364681
|
0.648
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr12_-_51200347
|
0.642
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr16_+_21623784
|
0.633
|
NM_170664
|
OTOA
|
otoancorin
|
|
chr12_-_60872814
|
0.633
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr2_-_29997935
|
0.629
|
NM_004304
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase
|
|
chr3_-_62836052
|
0.628
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr12_-_50971516
|
0.625
|
NM_002281
|
KRT81
|
keratin 81
|
|
chr6_+_47774247
|
0.620
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr20_+_43531806
|
0.610
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr9_-_34652629
|
0.604
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr5_-_147266257
|
0.600
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr4_-_54152480
|
0.599
|
NM_001126328
|
LNX1
|
ligand of numb-protein X 1
|
|
chr2_+_69055208
|
0.599
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr11_+_122214415
|
0.597
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr5_-_49772940
|
0.596
|
|
EMB
|
embigin
|
|
chr14_-_23106779
|
0.595
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr17_-_76919068
|
0.594
|
NM_178520
|
TMEM105
|
transmembrane protein 105
|
|
chr1_-_85703198
|
0.590
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr20_+_58064360
|
0.589
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr18_-_22696532
|
0.575
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr1_+_17404206
|
0.574
|
NM_013358
|
PADI1
|
peptidyl arginine deiminase, type I
|
|
chr20_-_1922701
|
0.571
|
NM_001190899
NM_001190898
NM_024411
|
PDYN
|
prodynorphin
|
|
chr15_-_88095363
|
0.566
|
NM_018670
|
MESP1
|
mesoderm posterior 1 homolog (mouse)
|
|
chr3_-_197549640
|
0.564
|
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr1_-_1131795
|
0.550
|
NM_004195
NM_148901
NM_148902
|
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18
|
|
chr5_-_39306465
|
0.549
|
|
FYB
|
FYN binding protein
|
|
chr5_-_1935764
|
0.548
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chrX_+_123307812
|
0.547
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr3_+_52786642
|
0.547
|
NM_002215
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr1_-_153098811
|
0.544
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr15_-_81271859
|
0.543
|
NM_001007122
|
FSD2
|
fibronectin type III and SPRY domain containing 2
|
|
chr2_-_218739891
|
0.540
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr6_+_31651319
|
0.540
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr14_+_51804180
|
0.538
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr3_-_156876798
|
0.538
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr1_-_159306369
|
0.537
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr4_-_140280031
|
0.536
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chrX_-_15783015
|
0.534
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr2_+_233112680
|
0.534
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chr1_+_154478574
|
0.532
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr3_-_186752952
|
0.523
|
NM_139248
|
LIPH
|
lipase, member H
|
|
chr12_-_8706746
|
0.521
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr1_+_154389945
|
0.521
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr12_-_8706533
|
0.517
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr6_-_159386007
|
0.516
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr16_-_28457728
|
0.512
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr10_+_24537725
|
0.511
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr17_+_18221803
|
0.510
|
NM_001145127
|
EVPLL
|
envoplakin-like
|
|
chr1_-_159306327
|
0.509
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr1_+_213245507
|
0.503
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr14_+_54104383
|
0.502
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr14_+_66725862
|
0.497
|
NM_173526
|
FAM71D
|
family with sequence similarity 71, member D
|
|
chr5_+_67622242
|
0.495
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_150819586
|
0.492
|
NM_032563
|
LCE3D
|
late cornified envelope 3D
|
|
chr1_+_149851112
|
0.492
|
|
|
|
|
chr16_-_80602546
|
0.490
|
NM_145168
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr11_-_128279670
|
0.483
|
|
C11orf45
|
chromosome 11 open reading frame 45
|
|
chr1_-_28392966
|
0.483
|
NM_001164722
NM_001164721
|
PTAFR
|
platelet-activating factor receptor
|
|
chr4_+_2596956
|
0.473
|
NM_003704
|
FAM193A
|
family with sequence similarity 193, member A
|
|
chr6_-_130578110
|
0.473
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr14_-_65550379
|
0.470
|
|
|
|
|
chrX_+_37750778
|
0.466
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr6_+_31059463
|
0.466
|
NM_001010909
|
MUC21
|
mucin 21, cell surface associated
|
|
chr1_+_201711505
|
0.460
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr12_+_10222823
|
0.450
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr2_+_27652892
|
0.446
|
NM_032266
|
C2orf16
|
chromosome 2 open reading frame 16
|
|
chr2_+_241213334
|
0.446
|
NM_005301
|
GPR35
|
G protein-coupled receptor 35
|
|
chr17_-_36559579
|
0.432
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr1_+_148496792
|
0.430
|
NM_012113
|
CA14
|
carbonic anhydrase XIV
|
|
chr14_-_23107106
|
0.429
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr5_-_132228375
|
0.427
|
NM_005260
|
GDF9
|
growth differentiation factor 9
|
|
chr5_+_156752739
|
0.424
|
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr2_+_218954995
|
0.423
|
NM_000578
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr12_+_55896844
|
0.422
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr1_+_180836050
|
0.419
|
|
|
|
|
chr2_+_131203112
|
0.419
|
NM_207364
|
GPR148
|
G protein-coupled receptor 148
|
|
chr11_+_34599163
|
0.418
|
NM_012153
|
EHF
|
ets homologous factor
|
|
chr20_+_32926452
|
0.418
|
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr11_-_65120042
|
0.413
|
NM_005714
NM_033347
NM_033348
NM_033455
|
KCNK7
|
potassium channel, subfamily K, member 7
|
|
chr8_-_22145462
|
0.410
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr7_+_129634935
|
0.409
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr1_-_54392030
|
0.408
|
NM_201546
|
CDCP2
|
CUB domain containing protein 2
|
|
chr6_+_88024152
|
0.407
|
|
ZNF292
|
zinc finger protein 292
|
|
chr1_-_159306210
|
0.406
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr2_+_165858586
|
0.405
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chrX_-_15782826
|
0.404
|
|
|
|
|
chr8_+_75899326
|
0.399
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr6_+_167456230
|
0.398
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|