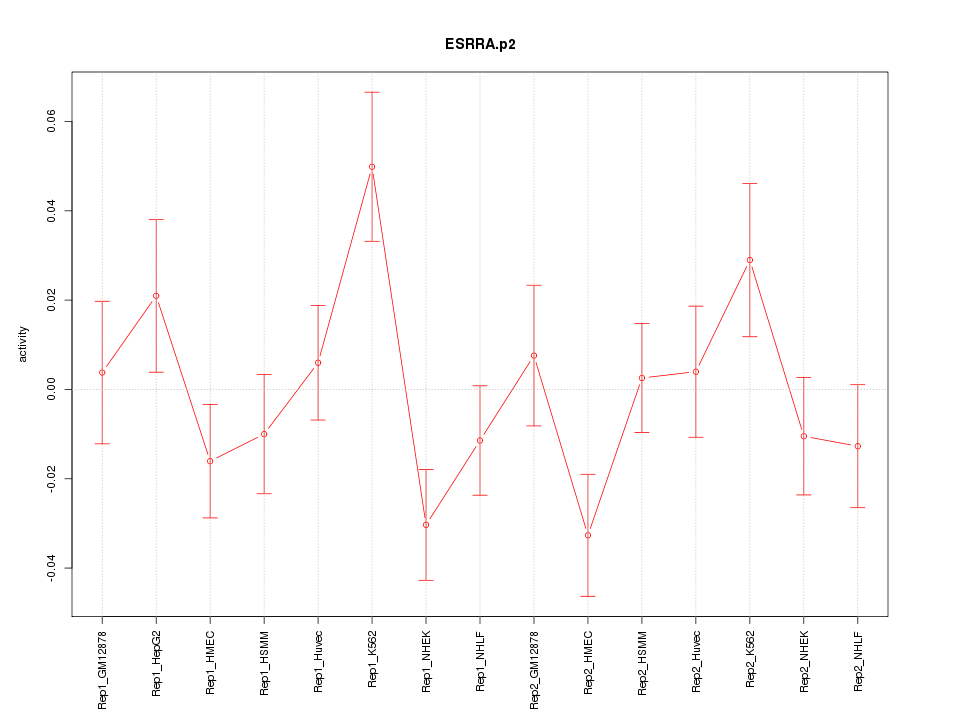
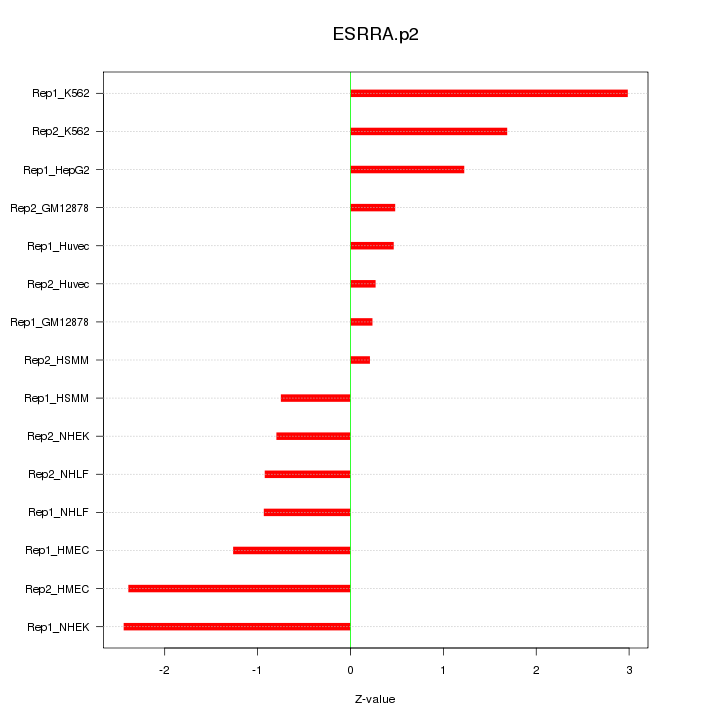
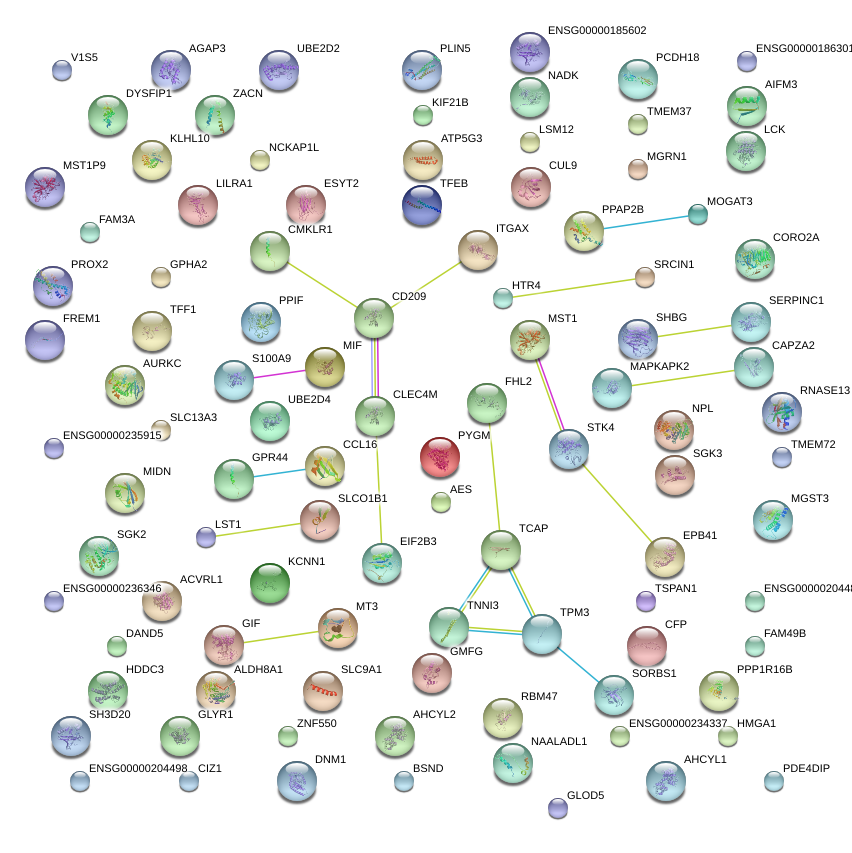
|
chr10_+_44726635
|
2.948
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr19_-_4486202
|
2.750
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr6_+_44076281
|
2.546
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr19_-_44518465
|
2.359
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr1_-_143706265
|
2.293
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_+_29086206
|
2.254
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr19_-_44518516
|
2.204
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr1_+_29086156
|
2.167
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr14_+_87560646
|
2.162
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr20_+_41621119
|
1.887
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr20_+_41621079
|
1.844
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr1_-_172153087
|
1.843
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr20_+_36867708
|
1.801
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr1_-_143706196
|
1.796
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr6_+_31662454
|
1.687
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr1_-_172153024
|
1.663
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr12_+_50592379
|
1.638
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr2_-_175754366
|
1.626
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr1_-_152431175
|
1.602
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr1_+_181028016
|
1.550
|
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr1_+_55237193
|
1.454
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr1_+_32489426
|
1.342
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr11_-_64284762
|
1.320
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr9_-_130006309
|
1.314
|
NM_001131015
NM_012127
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr3_-_49701085
|
1.295
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr17_-_39500512
|
1.273
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr17_+_37247568
|
1.268
|
NM_152467
|
KLHL10
|
kelch-like 10 (Drosophila)
|
|
chr19_+_7734034
|
1.266
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr11_-_60379998
|
1.243
|
NM_004778
|
GPR44
|
G protein-coupled receptor 44
|
|
chr19_+_62434188
|
1.178
|
NM_001015879
NM_003160
NM_001015878
|
AURKC
|
aurora kinase C
|
|
chr19_-_60360768
|
1.152
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr1_-_56750235
|
1.109
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_21631992
|
1.101
|
|
|
|
|
chr11_-_59369501
|
1.067
|
NM_005142
|
GIF
|
gastric intrinsic factor (vitamin B synthesis)
|
|
chr12_-_107257205
|
1.049
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr17_-_77386212
|
1.016
|
NM_001007533
|
DYSFIP1
|
dysferlin interacting protein 1
|
|
chr17_-_77386156
|
1.010
|
|
DYSFIP1
|
dysferlin interacting protein 1
|
|
chr19_+_17923110
|
0.981
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr1_-_35097922
|
0.971
|
NM_001164825
NM_138428
|
C1orf212
|
chromosome 1 open reading frame 212
|
|
chrX_-_153397566
|
0.965
|
NM_001171132
NM_001171133
NM_001171134
NM_021806
|
FAM3A
|
family with sequence similarity 3, member A
|
|
chrX_-_47374187
|
0.963
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr10_+_80777265
|
0.961
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr10_-_97190885
|
0.954
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr21_-_42659671
|
0.953
|
NM_003225
|
TFF1
|
trefoil factor 1
|
|
chr19_-_62759536
|
0.945
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr7_+_150413814
|
0.945
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr7_+_128795280
|
0.933
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr7_+_128795178
|
0.914
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr8_-_131097856
|
0.913
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr7_-_38355949
|
0.897
|
|
|
|
|
chr9_+_130005429
|
0.894
|
NM_001005336
NM_004408
|
DNM1
|
dynamin 1
|
|
chr6_-_135312899
|
0.893
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr6_+_34314545
|
0.891
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_-_3013880
|
0.886
|
NM_198969
|
AES
|
amino-terminal enhancer of split
|
|
chr1_-_199259447
|
0.884
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr9_+_130005489
|
0.868
|
|
DNM1
|
dynamin 1
|
|
chr17_-_31332568
|
0.860
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr17_+_35075124
|
0.859
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr15_-_89276657
|
0.857
|
NM_198527
|
HDDC3
|
HD domain containing 3
|
|
chr9_+_130005487
|
0.855
|
|
DNM1
|
dynamin 1
|
|
chr1_-_45224784
|
0.845
|
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr19_+_1199548
|
0.840
|
NM_177401
|
MIDN
|
midnolin
|
|
chr14_-_20572783
|
0.831
|
NM_001012264
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active)
|
|
chr4_-_40212670
|
0.831
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr10_+_80777248
|
0.823
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr7_-_158304062
|
0.820
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr7_+_116238309
|
0.813
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr9_-_14858974
|
0.806
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr22_+_19651446
|
0.805
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr11_-_64459910
|
0.797
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr7_-_38356100
|
0.785
|
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chrX_+_65152004
|
0.767
|
|
|
|
|
chr17_-_34014390
|
0.762
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr16_+_31274020
|
0.755
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chrX_+_48505097
|
0.754
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr16_-_4794749
|
0.750
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr8_-_131097828
|
0.742
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr6_+_43257883
|
0.734
|
NM_015089
|
CUL9
|
cullin 9
|
|
chr3_-_198839026
|
0.728
|
|
LOC220729
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene
|
|
chr14_-_74400288
|
0.724
|
NM_001080408
|
PROX2
|
prospero homeobox 2
|
|
chr16_+_31273969
|
0.722
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr19_-_3013768
|
0.711
|
|
AES
|
amino-terminal enhancer of split
|
|
chr16_+_4614959
|
0.708
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr7_-_100630912
|
0.708
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr1_-_27353987
|
0.700
|
NM_003047
|
SLC9A1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1
|
|
chr1_-_1679940
|
0.698
|
NM_001198995
|
NADK
|
NAD kinase
|
|
chr16_+_31274030
|
0.690
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr1_+_163867057
|
0.688
|
NM_004528
|
MGST3
|
microsomal glutathione S-transferase 3
|
|
chr6_+_44418374
|
0.676
|
NM_145026
|
SPATS1
|
spermatogenesis associated, serine-rich 1
|
|
chr5_-_148014282
|
0.675
|
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr6_-_41811323
|
0.674
|
|
TFEB
|
transcription factor EB
|
|
chr1_+_29113589
|
0.673
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_-_143706300
|
0.670
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_+_12941431
|
0.667
|
NM_152654
|
DAND5
|
DAN domain family, member 5
|
|
chr17_+_7474177
|
0.665
|
NM_001040
NM_001146279
NM_001146280
NM_001146281
|
SHBG
|
sex hormone-binding globulin
|
|
chr2_-_105421661
|
0.662
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr1_+_204924903
|
0.651
|
NM_004759
NM_032960
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2
|
|
chr5_+_138920893
|
0.646
|
NM_003339
NM_181838
|
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast)
|
|
chr19_+_59796858
|
0.646
|
NM_006863
|
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1
|
|
chrX_-_47374647
|
0.642
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr17_+_71586857
|
0.641
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr17_-_40839082
|
0.637
|
NM_001159330
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr20_-_44713466
|
0.637
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr9_-_99994704
|
0.632
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr2_+_119905812
|
0.631
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr6_-_31789824
|
0.630
|
|
|
|
|
chr11_-_64582568
|
0.630
|
NM_005468
|
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1
|
|
chr1_+_46413335
|
0.629
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr19_-_14771947
|
0.629
|
NM_198944
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1
|
|
chr6_-_108252200
|
0.627
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr4_+_24406336
|
0.615
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr3_+_46423682
|
0.607
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr1_+_226337425
|
0.605
|
NM_001024227
NM_001024228
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr6_+_36773605
|
0.603
|
|
RAB44
|
RAB44, member RAS oncogene family
|
|
chr16_-_65984836
|
0.597
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr12_+_56462794
|
0.589
|
NM_001172695
NM_001172696
NM_001172697
NM_005726
|
TSFM
|
Ts translation elongation factor, mitochondrial
|
|
chr13_-_79810972
|
0.588
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr4_+_24406238
|
0.583
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr21_+_43903803
|
0.580
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr3_+_187917791
|
0.578
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr4_+_24406301
|
0.576
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr4_+_24406182
|
0.574
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr11_-_18704294
|
0.573
|
NM_173588
|
IGSF22
|
immunoglobulin superfamily, member 22
|
|
chr6_+_31662954
|
0.567
|
NM_001166538
NM_007161
NM_205840
|
LST1
|
leukocyte specific transcript 1
|
|
chr18_-_44730706
|
0.565
|
|
SMAD7
|
SMAD family member 7
|
|
chr16_+_66121040
|
0.565
|
NM_001193523
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr20_+_31541565
|
0.564
|
NM_001032999
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr20_+_30061905
|
0.559
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr12_-_642786
|
0.557
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr1_-_150065176
|
0.556
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr1_-_38243747
|
0.552
|
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr22_-_29290875
|
0.552
|
NM_004861
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr4_+_24406283
|
0.547
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr10_+_80777234
|
0.546
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr11_+_268569
|
0.542
|
NM_138329
|
NLRP6
|
NLR family, pyrin domain containing 6
|
|
chr5_+_138921041
|
0.540
|
|
UBE2D2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast)
|
|
chr2_-_30883775
|
0.529
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr6_-_30236667
|
0.528
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr12_-_116021473
|
0.525
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr1_-_27689129
|
0.523
|
|
WASF2
|
WAS protein family, member 2
|
|
chr3_+_10265357
|
0.520
|
|
TATDN2
|
TatD DNase domain containing 2
|
|
chr9_+_102229283
|
0.519
|
NM_001198807
NM_080655
NM_001198805
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr16_-_28843955
|
0.517
|
NM_024816
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr9_-_37894331
|
0.512
|
NM_033412
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr16_+_55456619
|
0.511
|
NM_000339
NM_001126107
NM_001126108
|
SLC12A3
|
solute carrier family 12 (sodium/chloride transporters), member 3
|
|
chr10_+_80777199
|
0.507
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr5_-_148013908
|
0.505
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr2_-_32344192
|
0.502
|
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr11_-_780065
|
0.500
|
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr19_+_50141068
|
0.497
|
NM_000483
|
APOC2
|
apolipoprotein C-II
|
|
chr2_-_96667710
|
0.495
|
|
KIAA1310
|
KIAA1310
|
|
chr20_+_1132097
|
0.495
|
NM_001009612
|
C20orf202
|
chromosome 20 open reading frame 202
|
|
chr20_+_30329096
|
0.492
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr17_+_40580466
|
0.492
|
NM_006460
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr16_-_56886380
|
0.491
|
NM_001080492
|
PRSS54
|
protease, serine, 54
|
|
chr12_-_55042793
|
0.489
|
NM_001638
|
APOF
|
apolipoprotein F
|
|
chr22_+_18485145
|
0.485
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_-_19089971
|
0.484
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr1_-_27689254
|
0.483
|
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr1_-_203985932
|
0.477
|
|
|
|
|
chr19_-_56582997
|
0.477
|
NM_001161748
NM_030657
|
LIM2
|
lens intrinsic membrane protein 2, 19kDa
|
|
chr12_-_119038941
|
0.475
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr6_+_43257903
|
0.475
|
|
CUL9
|
cullin 9
|
|
chr6_+_37081387
|
0.474
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr15_+_41597097
|
0.470
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr8_-_131098015
|
0.468
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr2_-_96667787
|
0.468
|
|
KIAA1310
|
KIAA1310
|
|
chrX_-_149817720
|
0.465
|
|
CD99L2
|
CD99 molecule-like 2
|
|
chr11_+_77577505
|
0.463
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr7_-_64088848
|
0.461
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chrX_+_152682772
|
0.458
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr1_+_204924931
|
0.457
|
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2
|
|
chr16_+_24475129
|
0.456
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_-_27582368
|
0.455
|
NM_207397
|
CD164L2
|
CD164 sialomucin-like 2
|
|
chr19_+_1358750
|
0.455
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr7_+_128099581
|
0.451
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr1_+_24701886
|
0.448
|
NM_013441
|
RCAN3
|
RCAN family member 3
|
|
chr16_+_4614820
|
0.447
|
NM_001142289
NM_001142290
NM_001142291
NM_015246
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr6_-_11219932
|
0.446
|
NM_207582
|
ERVFRDE1
|
HERV-FRD provirus ancestral Env polyprotein
|
|
chr13_+_112349358
|
0.445
|
NM_207440
|
C13orf35
|
chromosome 13 open reading frame 35
|
|
chr6_+_10629550
|
0.443
|
NM_145649
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr12_-_24628348
|
0.441
|
|
C12orf67
|
chromosome 12 open reading frame 67
|
|
chr20_-_581808
|
0.440
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr15_+_76644882
|
0.440
|
NM_000745
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5
|
|
chr11_-_780103
|
0.439
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr18_+_53967544
|
0.438
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_+_37926687
|
0.437
|
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr1_+_157441133
|
0.436
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr2_-_172675873
|
0.435
|
|
DLX2
|
distal-less homeobox 2
|
|
chr10_-_75304225
|
0.435
|
NM_001222
NM_172169
NM_172171
NM_172173
NM_172170
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr19_+_1358664
|
0.429
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr12_-_119038934
|
0.427
|
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr19_+_19183763
|
0.426
|
NM_004386
|
NCAN
|
neurocan
|
|
chr7_-_92686773
|
0.425
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr17_-_70430839
|
0.424
|
NM_173477
|
USH1G
|
Usher syndrome 1G (autosomal recessive)
|
|
chr1_-_202726068
|
0.422
|
NM_002646
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr1_+_16242833
|
0.419
|
NM_000085
|
CLCNKB
|
chloride channel Kb
|
|
chr1_+_177261798
|
0.417
|
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr8_-_100975417
|
0.414
|
NM_004374
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr20_+_31541441
|
0.414
|
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|