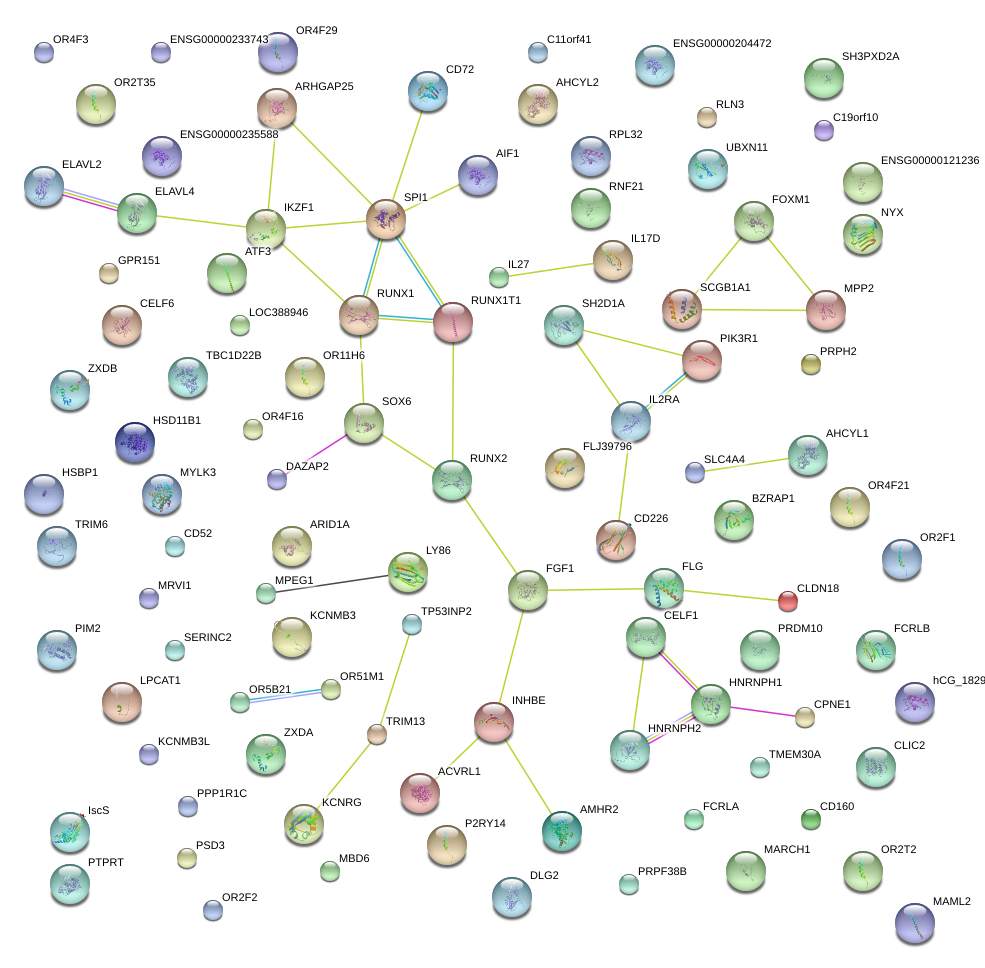
|
chr5_+_140871107
|
4.229
|
|
|
|
|
chr5_+_140870881
|
3.446
|
|
|
|
|
chr9_-_35608300
|
3.008
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr5_-_71838875
|
2.778
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr1_-_26517342
|
2.419
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr1_+_207944807
|
2.271
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr17_-_45626364
|
2.190
|
|
|
|
|
chr12_+_56206692
|
2.170
|
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chrX_+_41191630
|
2.169
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr17_-_53761119
|
2.158
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr11_+_59001421
|
2.056
|
NM_001004705
|
OR4D10
|
olfactory receptor, family 4, subfamily D, member 10
|
|
chr22_+_20715340
|
1.996
|
|
|
|
|
chr11_-_58032153
|
1.983
|
NM_001005218
|
OR5B21
|
olfactory receptor, family 5, subfamily B, member 21
|
|
chr1_+_31654998
|
1.948
|
NM_001199038
|
SERINC2
|
serine incorporator 2
|
|
chrX_-_154216929
|
1.934
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr11_+_33520348
|
1.908
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr7_+_50318801
|
1.887
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr18_-_65766068
|
1.831
|
|
CD226
|
CD226 molecule
|
|
chr4_+_72423633
|
1.769
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr16_-_28425632
|
1.704
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr5_-_178977441
|
1.666
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr15_-_70385709
|
1.643
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr20_+_32762976
|
1.637
|
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr3_+_139211639
|
1.622
|
NM_016369
|
CLDN18
|
claudin 18
|
|
chr12_+_52105131
|
1.612
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr15_-_75259714
|
1.553
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr19_-_16103996
|
1.551
|
|
|
|
|
chr3_-_180460372
|
1.543
|
NM_171829
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr11_-_47356562
|
1.542
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr5_-_145875868
|
1.540
|
NM_194251
|
GPR151
|
G protein-coupled receptor 151
|
|
chr16_-_45339605
|
1.529
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr11_+_5610060
|
1.513
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr2_+_46560207
|
1.478
|
NM_001145051
|
LOC388946
|
transmembrane protein ENSP00000343375
|
|
chr12_+_50592379
|
1.478
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr5_-_1547872
|
1.467
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr1_-_144426918
|
1.428
|
NM_007053
|
CD160
|
CD160 molecule
|
|
chr1_+_26516967
|
1.422
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr12_+_56135353
|
1.402
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr4_-_164754106
|
1.356
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr11_-_47356479
|
1.344
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr17_+_53761293
|
1.341
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr2_+_182558795
|
1.336
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr11_-_10630408
|
1.330
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr1_+_246682699
|
1.320
|
NM_001004136
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2
|
|
chr1_+_26517045
|
1.311
|
|
CD52
|
CD52 molecule
|
|
chr1_+_159959072
|
1.311
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr6_-_76022565
|
1.263
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr1_+_26973804
|
1.255
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr1_+_46910086
|
1.245
|
NM_001145474
|
C1orf223
|
chromosome 1 open reading frame 223
|
|
chr11_+_5367182
|
1.238
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chrX_-_48655978
|
1.232
|
|
PIM2
|
pim-2 oncogene
|
|
chrX_-_57953578
|
1.216
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr1_+_210805319
|
1.200
|
NM_001030287
|
ATF3
|
activating transcription factor 3
|
|
chr11_-_83071084
|
1.197
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_152478843
|
1.194
|
NM_014879
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr10_-_6144119
|
1.193
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr1_+_31655607
|
1.188
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr1_+_50348052
|
1.181
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_+_143263191
|
1.171
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chr3_-_12858006
|
1.165
|
NM_000994
NM_001007073
|
RPL32
|
ribosomal protein L32
|
|
chr11_-_58736897
|
1.162
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr5_+_67558217
|
1.158
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_-_95715976
|
1.151
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr5_-_141973875
|
1.144
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr10_-_105352098
|
1.140
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chrX_+_123307812
|
1.122
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr11_-_47356672
|
1.119
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr2_+_68815416
|
1.116
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr11_-_47356650
|
1.111
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr17_-_45626967
|
1.105
|
|
|
|
|
chr11_-_47356642
|
1.096
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr8_-_18585721
|
1.094
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_+_109043550
|
1.092
|
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr20_-_33705244
|
1.083
|
NM_003915
|
CPNE1
|
copine I
|
|
chr7_+_128795178
|
1.077
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_47467094
|
1.077
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr6_+_44076281
|
1.071
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr8_-_93176819
|
1.070
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_49922592
|
1.069
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr21_-_35153664
|
1.063
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr8_-_107023
|
1.059
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr7_+_128795280
|
1.059
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_-_129322510
|
1.051
|
NM_199438
NM_199439
|
PRDM10
|
PR domain containing 10
|
|
chr6_+_6533932
|
1.040
|
NM_004271
|
LY86
|
lymphocyte antigen 86
|
|
chr6_+_31691745
|
1.033
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr11_+_61943082
|
1.033
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr11_-_16380967
|
1.033
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr13_+_49487390
|
1.024
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr19_+_13999959
|
1.017
|
NM_080864
|
RLN3
|
relaxin 3
|
|
chr6_+_37360144
|
1.012
|
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr1_-_611896
|
1.011
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr14_+_19761708
|
1.005
|
NM_001004480
|
OR11H6
|
olfactory receptor, family 11, subfamily H, member 6
|
|
chr2_-_29150630
|
1.005
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr1_+_159899528
|
1.004
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr11_-_5110383
|
1.004
|
NM_001005160
|
OR52A5
|
olfactory receptor, family 52, subfamily A, member 5
|
|
chr5_+_180726874
|
0.997
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chrX_+_77267477
|
0.991
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr11_-_5178505
|
0.989
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr22_+_21484240
|
0.981
|
|
|
|
|
chr22_+_20880159
|
0.978
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr2_+_134728299
|
0.977
|
NM_002410
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr11_-_5523328
|
0.976
|
NM_001005289
|
OR52H1
|
olfactory receptor, family 52, subfamily H, member 1
|
|
chr11_-_5227611
|
0.970
|
NM_000559
|
HBG1
HBG2
|
hemoglobin, gamma A
hemoglobin, gamma G
|
|
chr16_+_3345889
|
0.957
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr16_+_142853
|
0.954
|
NM_005332
|
HBZ
|
hemoglobin, zeta
|
|
chr3_+_153499883
|
0.954
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chrX_-_133620103
|
0.950
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr1_-_180621272
|
0.948
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr22_-_35875890
|
0.940
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr16_-_87570716
|
0.933
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chrX_-_100527799
|
0.933
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr2_+_68855436
|
0.932
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr7_+_69869099
|
0.932
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr1_+_109450116
|
0.928
|
|
|
|
|
chr15_+_89570333
|
0.925
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr1_+_153232685
|
0.918
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr6_+_33151703
|
0.917
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr14_+_63752424
|
0.917
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr11_-_5487728
|
0.914
|
NM_017481
|
UBQLN3
|
ubiquilin 3
|
|
chr11_+_48241988
|
0.914
|
NM_001004726
|
OR4X1
|
olfactory receptor, family 4, subfamily X, member 1
|
|
chr3_+_52788976
|
0.903
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr2_-_74634569
|
0.901
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr1_+_109043711
|
0.899
|
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr3_+_99733432
|
0.897
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr1_-_120285753
|
0.896
|
|
NOTCH2
|
notch 2
|
|
chr2_-_97641489
|
0.893
|
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chrX_+_110252960
|
0.887
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr22_+_20880112
|
0.885
|
|
|
|
|
chr16_+_29582480
|
0.882
|
|
SPN
|
sialophorin
|
|
chrX_-_77469635
|
0.882
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr9_+_114376709
|
0.880
|
|
KIAA1958
|
KIAA1958
|
|
chr4_+_38740246
|
0.872
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr11_-_6148131
|
0.866
|
NM_001004052
|
OR52B2
|
olfactory receptor, family 52, subfamily B, member 2
|
|
chr1_+_26517032
|
0.865
|
|
CD52
|
CD52 molecule
|
|
chr1_-_160648429
|
0.864
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr1_+_357502
|
0.854
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr1_-_246869181
|
0.848
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr9_-_122728173
|
0.843
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr6_-_76259976
|
0.841
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr6_-_32259974
|
0.838
|
NM_001136
NM_172197
|
AGER
|
advanced glycosylation end product-specific receptor
|
|
chr15_+_57286313
|
0.835
|
NM_033195
|
LDHAL6B
|
lactate dehydrogenase A-like 6B
|
|
chr1_-_239865724
|
0.831
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chrX_-_55037235
|
0.829
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr14_-_23117614
|
0.828
|
|
JPH4
|
junctophilin 4
|
|
chr9_+_123126652
|
0.828
|
|
GSN
|
gelsolin
|
|
chr10_+_85923473
|
0.827
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr6_+_52159143
|
0.826
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chr13_-_45640679
|
0.822
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr4_-_123073642
|
0.822
|
NM_003305
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr18_+_41560915
|
0.821
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr19_+_6723801
|
0.813
|
|
|
|
|
chr15_-_41879426
|
0.813
|
NM_001033517
|
SERINC4
|
serine incorporator 4
|
|
chr9_-_99740243
|
0.813
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr11_-_64403712
|
0.812
|
|
EHD1
|
EH-domain containing 1
|
|
chr16_-_46738061
|
0.811
|
NM_033226
|
ABCC12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12
|
|
chr19_+_6723705
|
0.805
|
NM_005428
|
VAV1
|
vav 1 guanine nucleotide exchange factor
|
|
chr12_+_1608672
|
0.804
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr1_+_154120170
|
0.802
|
|
SYT11
|
synaptotagmin XI
|
|
chr16_+_27346079
|
0.800
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr19_+_54669629
|
0.798
|
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr10_-_75080784
|
0.798
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr6_-_29120930
|
0.793
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chr17_-_23965305
|
0.787
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr10_-_105427898
|
0.787
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr12_-_66839589
|
0.784
|
NM_000619
|
IFNG
|
interferon, gamma
|
|
chrX_-_70243362
|
0.783
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr1_+_58899
|
0.780
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr12_+_5473558
|
0.779
|
NM_002527
|
NTF3
|
neurotrophin 3
|
|
chr15_-_42756377
|
0.777
|
NM_001145112
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast)
|
|
chr22_+_21359175
|
0.774
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr1_+_46745254
|
0.769
|
NM_147192
NM_172225
|
DMBX1
|
diencephalon/mesencephalon homeobox 1
|
|
chr17_+_19917420
|
0.767
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr16_-_66075216
|
0.765
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chr1_-_51660328
|
0.762
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr16_+_29892714
|
0.762
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr11_+_58967192
|
0.762
|
NM_001004728
|
OR5A1
|
olfactory receptor, family 5, subfamily A, member 1
|
|
chr17_-_1195096
|
0.760
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr19_+_54299686
|
0.758
|
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1)
|
|
chr9_+_124526089
|
0.755
|
NM_001005235
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4
|
|
chr3_-_120861959
|
0.753
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr12_+_48630790
|
0.752
|
NM_000486
|
AQP2
|
aquaporin 2 (collecting duct)
|
|
chr9_-_124280025
|
0.748
|
NM_001004451
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1
|
|
chr4_-_164614335
|
0.745
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr22_-_37211467
|
0.740
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr11_+_59038961
|
0.739
|
NM_001004711
|
OR4D9
|
olfactory receptor, family 4, subfamily D, member 9
|
|
chr20_-_34877972
|
0.730
|
|
KIAA0889
|
KIAA0889
|
|
chr6_+_7524691
|
0.729
|
|
DSP
|
desmoplakin
|
|
chr8_-_93099040
|
0.726
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_+_91244563
|
0.724
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr19_-_59495949
|
0.723
|
NM_001172654
NM_006865
|
LILRB2
LILRA3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr20_+_42007949
|
0.722
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr11_-_59369501
|
0.719
|
NM_005142
|
GIF
|
gastric intrinsic factor (vitamin B synthesis)
|
|
chr3_+_142587973
|
0.718
|
|
|
|
|
chr11_-_72057755
|
0.717
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr2_-_172082477
|
0.711
|
|
|
|
|
chr19_-_14085979
|
0.710
|
NM_207518
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr17_-_7047285
|
0.709
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr17_-_64568641
|
0.708
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr10_+_35524060
|
0.707
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr5_-_179040580
|
0.706
|
NM_001164444
|
CBY3
|
chibby homolog 3 (Drosophila)
|