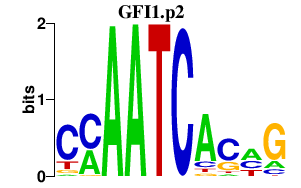
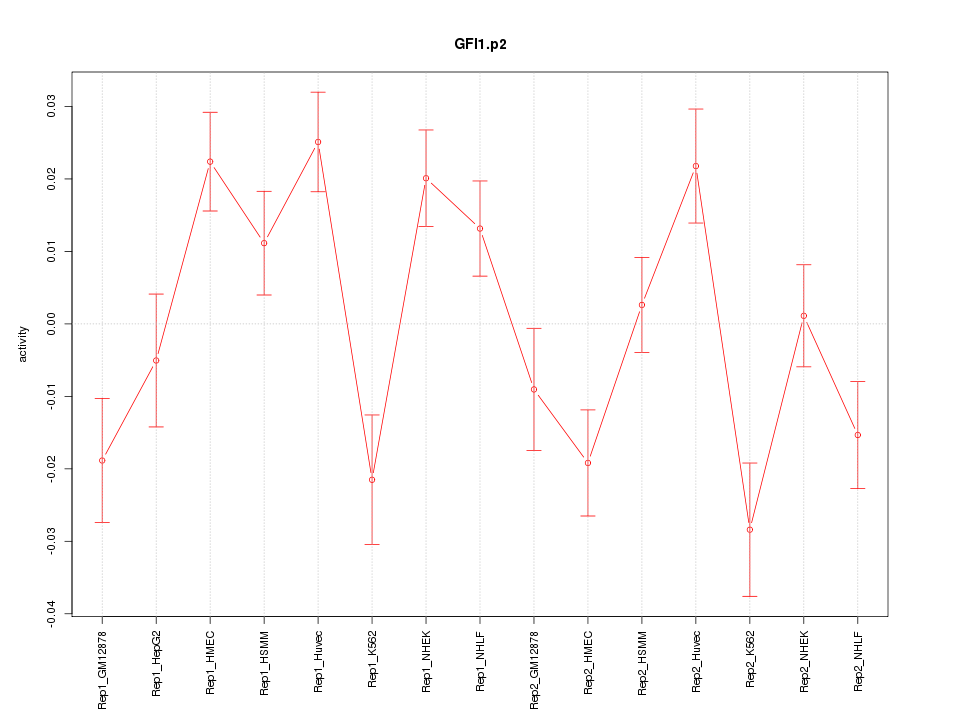
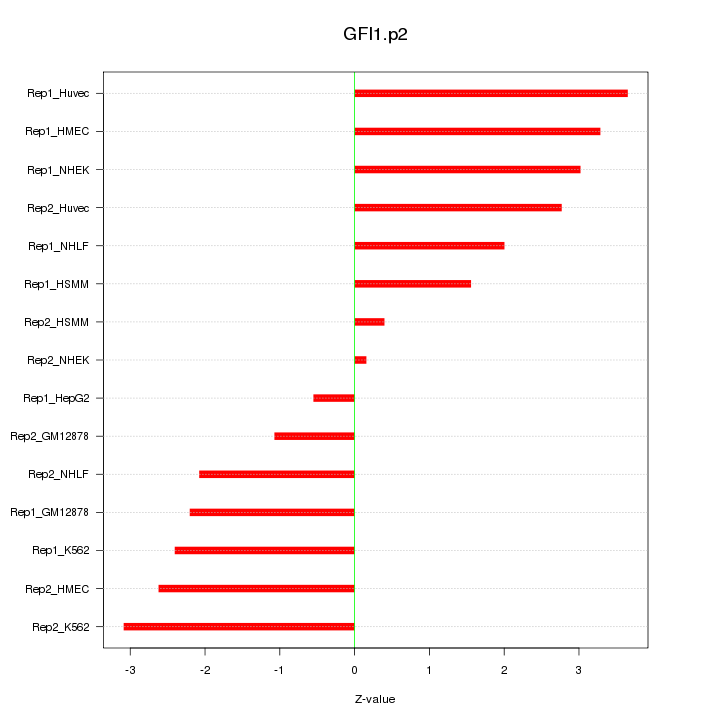
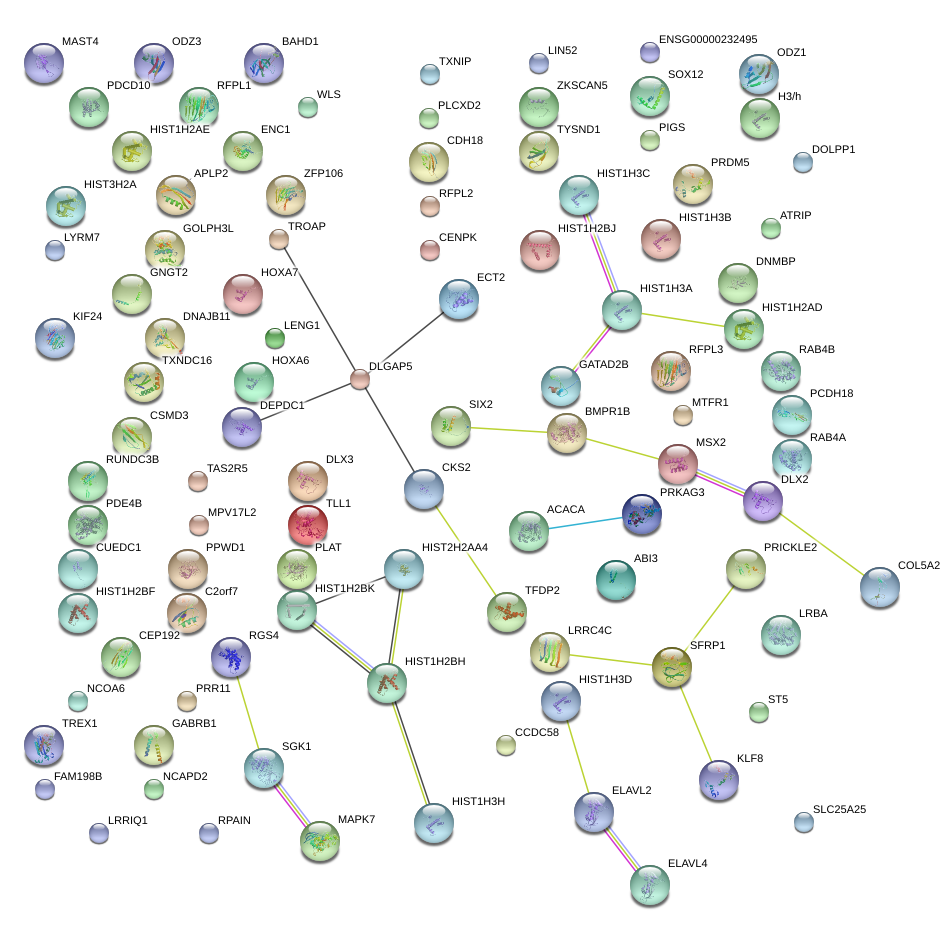
|
chr10_-_101680645
|
3.307
|
|
DNMBP
|
dynamin binding protein
|
|
chr7_-_27162688
|
2.640
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr9_-_23816062
|
2.624
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr5_+_66160359
|
2.398
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr6_+_26153590
|
1.895
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr9_+_130883199
|
1.732
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr3_-_168935287
|
1.716
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr4_+_167013859
|
1.706
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr9_-_34319151
|
1.661
|
NM_194313
|
KIF24
|
kinesin family member 24
|
|
chr3_+_173951163
|
1.660
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr22_+_31080871
|
1.655
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr17_+_44643067
|
1.647
|
|
ABI3
|
ABI family, member 3
|
|
chr3_+_112876212
|
1.614
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr8_-_41286088
|
1.542
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr14_-_54727958
|
1.479
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr19_+_18164989
|
1.469
|
NM_032683
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2
|
|
chr5_-_64894687
|
1.453
|
NM_022145
|
CENPK
|
centromere protein K
|
|
chr1_-_148936123
|
1.426
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr6_-_26140266
|
1.379
|
NM_003537
|
HIST1H3B
|
histone cluster 1, H3b
|
|
chr2_-_219404755
|
1.374
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr17_-_44642934
|
1.360
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr6_-_26379567
|
1.321
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr12_+_48003237
|
1.317
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr15_+_38520594
|
1.308
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr11_+_129496792
|
1.298
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr4_+_183302105
|
1.296
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr7_-_27153892
|
1.277
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr8_-_114518194
|
1.268
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr3_-_64186151
|
1.262
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr17_+_5264195
|
1.258
|
|
RPAIN
|
RPA interacting protein
|
|
chr14_+_73621408
|
1.244
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr7_+_87095640
|
1.224
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr1_-_152162074
|
1.219
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr5_+_64894854
|
1.208
|
NM_015342
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr6_-_27208507
|
1.201
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr4_-_151990061
|
1.181
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr19_+_45976001
|
1.174
|
NM_016154
|
RAB4B-EGLN2
RAB4B
|
RAB4B-EGLN2 read-through transcript
RAB4B, member RAS oncogene family
|
|
chr4_-_138673078
|
1.153
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_68470585
|
1.152
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr18_+_12981329
|
1.142
|
NM_032142
|
CEP192
|
centrosomal protein 192kDa
|
|
chr7_+_98940801
|
1.138
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr4_+_46728051
|
1.137
|
NM_000812
|
GABRB1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1
|
|
chr3_-_123584747
|
1.137
|
NM_001017928
|
CCDC58
|
coiled-coil domain containing 58
|
|
chrX_+_56275510
|
1.135
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr5_+_130534555
|
1.128
|
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr19_-_59355246
|
1.108
|
NM_024316
|
LENG1
|
leukocyte receptor cluster (LRC) member 1
|
|
chr17_-_45427565
|
1.108
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr4_-_138673014
|
1.104
|
|
PCDH18
|
protocadherin 18
|
|
chr12_+_6473552
|
1.078
|
NM_014865
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr3_-_143350898
|
1.069
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr12_+_6473519
|
1.067
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr10_+_102123589
|
1.064
|
|
NCRNA00263
|
non-protein coding RNA 263
|
|
chr15_-_40537002
|
1.063
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr1_-_68735267
|
1.058
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr6_-_26305456
|
1.049
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr4_-_159313613
|
1.040
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_-_53335748
|
1.040
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr11_-_8849485
|
1.033
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_+_161305558
|
1.026
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_144149794
|
1.026
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr20_+_254205
|
1.026
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr14_-_52088786
|
1.024
|
NM_001160047
NM_020784
|
TXNDC16
|
thioredoxin domain containing 16
|
|
chr2_-_45090025
|
1.020
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr17_+_54587882
|
0.999
|
|
PRR11
|
proline rich 11
|
|
chr2_-_189752575
|
0.999
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr5_+_174084137
|
0.990
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr9_+_129870268
|
0.990
|
NM_001006641
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr4_+_95898098
|
0.981
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr5_-_20024050
|
0.978
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr17_+_19221606
|
0.973
|
NM_139032
NM_139033
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr11_-_40272239
|
0.971
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr7_+_141136485
|
0.971
|
NM_018980
|
TAS2R5
|
taste receptor, type 2, member 5
|
|
chr8_-_42184350
|
0.970
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr3_+_48463184
|
0.958
|
NM_032166
NM_130384
|
TREX1
ATRIP
|
three prime repair exonuclease 1
ATR interacting protein
|
|
chr5_-_73973004
|
0.957
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr3_+_187771192
|
0.955
|
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr12_+_83954229
|
0.952
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr8_+_66719441
|
0.949
|
NM_001145839
NM_014637
|
MTFR1
|
mitochondrial fission regulator 1
|
|
chr8_-_93144039
|
0.948
|
|
|
|
|
chr9_+_129870416
|
0.948
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr4_-_122063118
|
0.939
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr17_-_32730804
|
0.938
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr12_-_120725210
|
0.936
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr6_-_134540666
|
0.934
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_-_23922622
|
0.929
|
|
PIGS
|
phosphatidylinositol glycan anchor biosynthesis, class S
|
|
chr2_-_73313823
|
0.927
|
NM_032319
|
C2orf7
|
chromosome 2 open reading frame 7
|
|
chr9_+_91115924
|
0.922
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr1_+_66570683
|
0.922
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr10_-_71576458
|
0.919
|
NM_001040273
NM_173555
|
TYSND1
|
trypsin domain containing 1
|
|
chr12_+_49728350
|
0.919
|
NM_001024668
NM_015416
|
LETMD1
|
LETM1 domain containing 1
|
|
chr3_+_129355006
|
0.917
|
|
EEFSEC
|
eukaryotic elongation factor, selenocysteine-tRNA-specific
|
|
chr1_-_109957195
|
0.917
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr20_-_54400628
|
0.917
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr19_-_11352850
|
0.913
|
|
EPOR
|
erythropoietin receptor
|
|
chr17_+_20970828
|
0.911
|
NM_015510
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B
|
|
chr5_+_57914658
|
0.911
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr3_+_132583204
|
0.907
|
NM_001171905
NM_001171906
NM_152395
|
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr22_+_40195058
|
0.903
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr4_-_76774594
|
0.900
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr5_-_59819642
|
0.897
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_66570363
|
0.897
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr12_-_101037976
|
0.889
|
|
NUP37
|
nucleoporin 37kDa
|
|
chr3_-_116348780
|
0.889
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_+_115957537
|
0.884
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr6_+_26333361
|
0.879
|
NM_003532
|
HIST1H3E
|
histone cluster 1, H3e
|
|
chr17_-_37983175
|
0.878
|
NM_013290
NM_016556
|
PSMC3IP
|
PSMC3 interacting protein
|
|
chr5_+_102229612
|
0.876
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr11_+_13255836
|
0.874
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr2_-_111152075
|
0.872
|
NM_004336
|
BUB1
|
budding uninhibited by benzimidazoles 1 homolog (yeast)
|
|
chr11_-_70837090
|
0.866
|
NM_001163817
NM_001360
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr6_-_26125939
|
0.864
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr16_+_81218074
|
0.862
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr5_+_130534534
|
0.860
|
NM_181705
|
LYRM7
|
Lyrm7 homolog (mouse)
|
|
chr3_-_198948110
|
0.856
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr2_+_219180821
|
0.851
|
|
PLCD4
|
phospholipase C, delta 4
|
|
chr9_-_94095810
|
0.847
|
NM_002161
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr13_-_77391525
|
0.843
|
|
|
|
|
chr15_+_98960277
|
0.841
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr16_-_55777376
|
0.841
|
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr11_-_47154955
|
0.841
|
NM_032389
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr9_+_676640
|
0.840
|
|
|
|
|
chr4_-_5761194
|
0.840
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr6_+_27885692
|
0.836
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr9_-_94095807
|
0.834
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr9_-_94095787
|
0.833
|
NM_013417
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr10_-_123343320
|
0.827
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr21_+_29439768
|
0.827
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr19_-_9982041
|
0.825
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr1_-_68735314
|
0.824
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr5_-_100266769
|
0.823
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr17_+_56109940
|
0.820
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr3_+_187771139
|
0.819
|
NM_016306
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11
|
|
chr6_+_32970477
|
0.819
|
|
LOC100294145
|
hypothetical LOC100294145
|
|
chr6_+_36752244
|
0.816
|
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr11_+_58631190
|
0.813
|
NM_001142703
NM_001142704
NM_198947
|
FAM111B
|
family with sequence similarity 111, member B
|
|
chr3_-_69517913
|
0.806
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr10_-_101180191
|
0.798
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr17_-_5330204
|
0.794
|
NM_016041
|
DERL2
|
Der1-like domain family, member 2
|
|
chr11_-_102100833
|
0.786
|
NM_002424
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase)
|
|
chr5_-_108773468
|
0.784
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr16_-_76026431
|
0.782
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr17_-_57295451
|
0.780
|
NM_032043
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr11_-_117640407
|
0.777
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr6_+_27941012
|
0.776
|
NM_003511
|
HIST1H2AI
HIST1H2AL
|
histone cluster 1, H2ai
histone cluster 1, H2al
|
|
chr3_+_185537969
|
0.772
|
NM_144635
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr9_-_90983501
|
0.772
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr2_+_219180875
|
0.770
|
NM_032726
|
PLCD4
|
phospholipase C, delta 4
|
|
chr10_+_105871805
|
0.770
|
NM_145247
NM_001002759
|
MEIR5
|
MEI5 meiotic recombination protein homolog (S. cerevisiae)
|
|
chr9_-_94095734
|
0.769
|
|
IARS
|
isoleucyl-tRNA synthetase
|
|
chr5_+_102229425
|
0.769
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr6_+_26307691
|
0.764
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr5_+_64956586
|
0.763
|
|
C5orf44
|
chromosome 5 open reading frame 44
|
|
chr14_+_73388259
|
0.762
|
NM_001146154
NM_001146155
NM_152444
|
PTGR2
|
prostaglandin reductase 2
|
|
chr3_-_187771025
|
0.760
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr11_-_62355128
|
0.760
|
|
STX5
|
syntaxin 5
|
|
chr2_-_216944911
|
0.754
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr4_+_86968069
|
0.752
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_-_127901633
|
0.750
|
NM_001999
|
FBN2
|
fibrillin 2
|
|
chr17_+_73694992
|
0.750
|
NM_001010982
NM_001145526
|
AFMID
|
arylformamidase
|
|
chr10_-_123346148
|
0.749
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr1_+_161305188
|
0.749
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_83954185
|
0.747
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr9_+_136673534
|
0.746
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr5_+_162797149
|
0.745
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr14_+_51526071
|
0.740
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr6_-_109521969
|
0.738
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr4_-_118226119
|
0.736
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr17_+_44642587
|
0.735
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr10_-_104463947
|
0.731
|
NM_004311
|
ARL3
|
ADP-ribosylation factor-like 3
|
|
chr3_+_114414064
|
0.729
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr3_+_170974745
|
0.726
|
NM_001185119
|
MYNN
|
myoneurin
|
|
chr1_-_147665783
|
0.721
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr15_+_57517564
|
0.720
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr4_-_89963177
|
0.719
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr12_-_47532171
|
0.718
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr3_-_123036526
|
0.718
|
|
IQCB1
|
IQ motif containing B1
|
|
chr15_+_67532176
|
0.716
|
NM_001003
NM_213725
|
RPLP1
|
ribosomal protein, large, P1
|
|
chr19_-_63643400
|
0.713
|
NM_003433
|
ZNF132
|
zinc finger protein 132
|
|
chr2_+_85676347
|
0.712
|
NM_016494
|
RNF181
|
ring finger protein 181
|
|
chr14_-_72430545
|
0.711
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr12_+_111829121
|
0.704
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr12_-_8993515
|
0.702
|
NM_002355
|
M6PR
|
mannose-6-phosphate receptor (cation dependent)
|
|
chr17_+_31160550
|
0.701
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr1_+_112740322
|
0.701
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr11_+_71317415
|
0.700
|
NM_018320
|
RNF121
|
ring finger protein 121
|
|
chr3_+_168936125
|
0.696
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr6_+_27883877
|
0.694
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr14_+_90650720
|
0.691
|
NM_001102367
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr17_+_19222651
|
0.690
|
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr12_+_121577733
|
0.688
|
NM_014708
|
KNTC1
|
kinetochore associated 1
|
|
chr15_-_71712711
|
0.685
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr5_-_110876186
|
0.681
|
|
STARD4
|
StAR-related lipid transfer (START) domain containing 4
|
|
chr2_+_219180862
|
0.677
|
|
PLCD4
|
phospholipase C, delta 4
|
|
chrX_-_33139349
|
0.675
|
NM_004006
|
DMD
|
dystrophin
|
|
chr6_+_27969181
|
0.674
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr3_-_53139457
|
0.672
|
NM_052859
|
RFT1
|
RFT1 homolog (S. cerevisiae)
|
|
chr9_+_136673464
|
0.669
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr12_+_122283796
|
0.667
|
NM_152269
NM_001194995
|
C12orf65
|
chromosome 12 open reading frame 65
|
|
chr18_+_3402071
|
0.667
|
NM_174886
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr5_+_15553288
|
0.667
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|