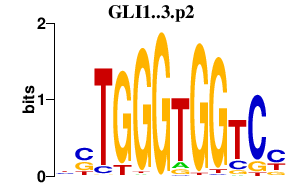
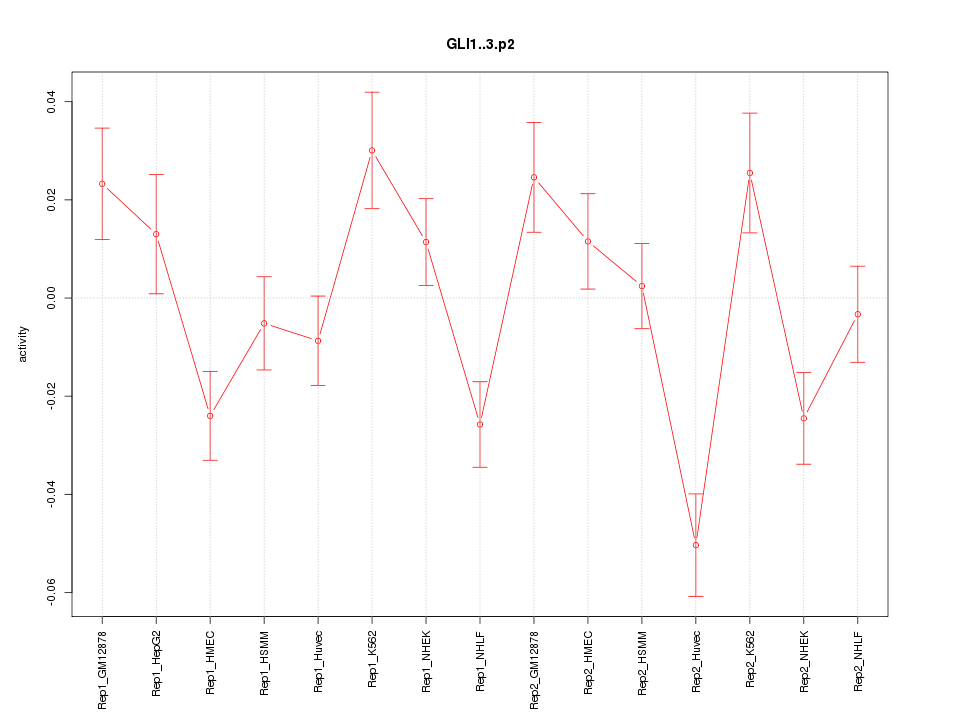
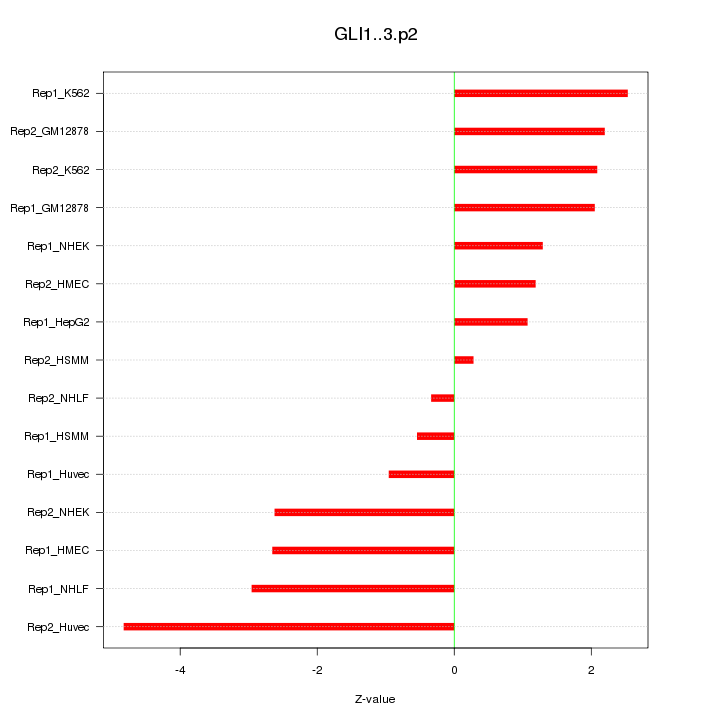
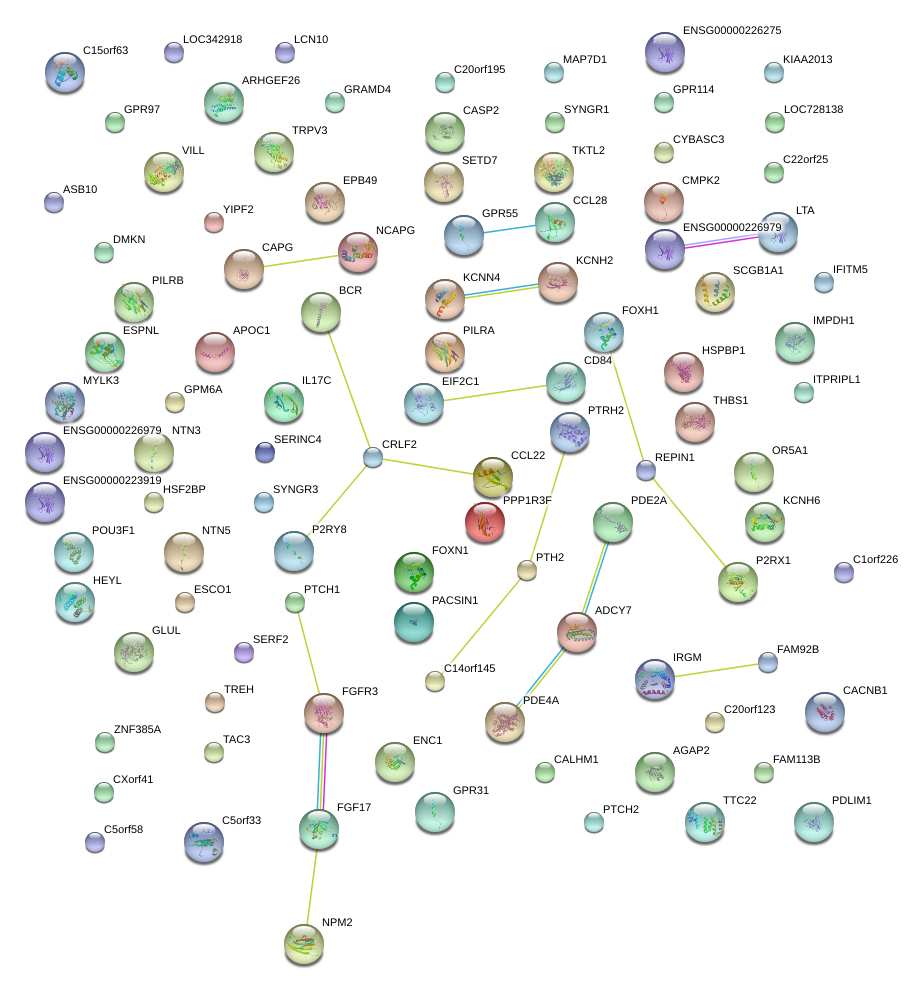
|
chr11_-_72057755
|
3.633
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr11_-_60880799
|
3.540
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr22_+_18090118
|
3.175
|
|
SEPT5-GP1BB
|
SEPT5-GP1BB read-through transcript
|
|
chr16_+_48865521
|
3.077
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr20_-_44612619
|
2.515
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr16_+_56259657
|
2.414
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr8_+_21972631
|
2.403
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr6_+_44076281
|
2.129
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr18_-_17434811
|
1.954
|
|
ESCO1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr20_-_42457717
|
1.952
|
|
|
|
|
chr17_+_73738985
|
1.946
|
NM_001145529
|
TMEM235
|
transmembrane protein 235
|
|
chr9_+_130056513
|
1.889
|
|
|
|
|
chr8_+_21972804
|
1.839
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr1_-_11909042
|
1.775
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr16_+_2461500
|
1.773
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr17_-_3407780
|
1.726
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr1_-_180621272
|
1.680
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr19_+_55844446
|
1.604
|
NM_001195076
|
LOC342918
|
hypothetical protein LOC342918
|
|
chr19_-_48977247
|
1.599
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr17_-_3408038
|
1.577
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr17_-_3766441
|
1.547
|
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1
|
|
chr19_+_2240996
|
1.480
|
|
|
|
|
chr2_-_231498184
|
1.417
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr11_-_289518
|
1.407
|
NM_001025295
|
IFITM5
|
interferon induced transmembrane protein 5
|
|
chr12_-_53064723
|
1.372
|
NM_015481
|
ZNF385A
|
zinc finger protein 385A
|
|
chr5_-_43448174
|
1.351
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr17_-_3766708
|
1.327
|
NM_002558
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1
|
|
chr10_-_105208634
|
1.324
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr4_-_140696740
|
1.301
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr7_-_142695392
|
1.301
|
|
|
|
|
chr17_+_23875085
|
1.288
|
NM_003593
|
FOXN1
|
forkhead box N1
|
|
chr8_+_21937533
|
1.283
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr7_-_150515851
|
1.259
|
NM_080871
|
ASB10
|
ankyrin repeat and SOCS box containing 10
|
|
chr15_-_41879426
|
1.212
|
NM_001033517
|
SERINC4
|
serine incorporator 4
|
|
chr15_+_37660568
|
1.208
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr4_-_164614335
|
1.205
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr8_-_145671947
|
1.194
|
|
FOXH1
|
forkhead box H1
|
|
chr9_-_97319067
|
1.189
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr9_-_138757175
|
1.184
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr19_+_50109416
|
1.178
|
|
APOC1
|
apolipoprotein C-I
|
|
chr16_-_45339605
|
1.168
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr9_-_97318760
|
1.158
|
|
PTCH1
|
patched 1
|
|
chr19_-_54618410
|
1.149
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chr7_+_142695570
|
1.140
|
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr7_-_150306169
|
1.128
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr1_-_45081190
|
1.112
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr19_+_10404110
|
1.098
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr3_+_38010081
|
1.093
|
NM_015873
|
VILL
|
villin-like
|
|
chr12_-_53064565
|
1.089
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr11_+_61943082
|
1.078
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr1_+_160615319
|
1.065
|
NM_001135240
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr5_-_168204430
|
1.056
|
|
|
|
|
chr4_-_176970895
|
1.055
|
|
GPM6A
|
glycoprotein M6A
|
|
chr20_+_61655934
|
1.055
|
|
|
|
|
chr20_+_61654816
|
1.053
|
NM_024059
|
C20orf195
|
chromosome 20 open reading frame 195
|
|
chr1_-_158815850
|
1.053
|
|
CD84
|
CD84 molecule
|
|
chr16_+_87232501
|
1.050
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr11_-_118055564
|
1.050
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr16_+_56134077
|
1.049
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr16_-_83703496
|
1.044
|
NM_198491
|
FAM92B
|
family with sequence similarity 92, member B
|
|
chr7_+_149699185
|
1.036
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr7_+_142695511
|
1.023
|
NM_032982
NM_032983
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr10_-_97040661
|
1.022
|
NM_020992
|
PDLIM1
|
PDZ and LIM domain 1
|
|
chr12_+_45896511
|
0.994
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr19_-_60483302
|
0.993
|
NM_001130106
NM_012267
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr12_-_56422120
|
0.992
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr16_+_56134037
|
0.988
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr7_-_127833231
|
0.985
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr1_-_38285033
|
0.979
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr22_+_38090120
|
0.974
|
NM_145738
|
SYNGR1
|
synaptogyrin 1
|
|
chr1_-_39877928
|
0.971
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr16_+_55950195
|
0.954
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr2_-_85490984
|
0.948
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr18_-_17434655
|
0.947
|
NM_052911
|
ESCO1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr1_-_55039458
|
0.947
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr2_+_238673689
|
0.926
|
NM_194312
|
ESPNL
|
espin-like
|
|
chr19_-_10900086
|
0.912
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr1_+_199259664
|
0.911
|
|
|
|
|
chr8_+_21956335
|
0.903
|
NM_003867
|
FGF17
|
fibroblast growth factor 17
|
|
chr6_+_31647854
|
0.903
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr12_-_55696542
|
0.896
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr17_-_34607257
|
0.891
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr22_+_45401321
|
0.889
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr22_+_18388633
|
0.887
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr2_-_6923215
|
0.879
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr22_+_20899155
|
0.875
|
|
|
|
|
chr19_+_50109903
|
0.869
|
|
APOC1
|
apolipoprotein C-I
|
|
chr19_-_48976852
|
0.862
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_-_39877883
|
0.861
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr6_+_34541827
|
0.852
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr1_+_36394044
|
0.850
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr19_-_53868075
|
0.849
|
NM_145807
|
NTN5
|
netrin 5
|
|
chr2_+_96355661
|
0.849
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr5_-_36277656
|
0.844
|
NM_001085411
|
C5orf33
|
chromosome 5 open reading frame 33
|
|
chr5_+_150206277
|
0.843
|
NM_001145805
|
IRGM
|
immunity-related GTPase family, M
|
|
chr6_-_167491305
|
0.842
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr14_-_106352471
|
0.841
|
|
|
|
|
chr19_+_10388448
|
0.841
|
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr15_+_41879912
|
0.836
|
NM_016400
|
C15orf63
|
chromosome 15 open reading frame 63
|
|
chr19_-_40684638
|
0.834
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr16_+_1979959
|
0.833
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chrX_+_106336517
|
0.831
|
NM_001169154
NM_173494
|
CXorf41
|
chromosome X open reading frame 41
|
|
chr1_+_36121382
|
0.829
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr22_+_21852911
|
0.828
|
|
BCR
|
breakpoint cluster region
|
|
chr7_-_150305933
|
0.826
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr11_+_58967192
|
0.825
|
NM_001004728
|
OR5A1
|
olfactory receptor, family 5, subfamily A, member 1
|
|
chr4_+_1764802
|
0.825
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chrX_-_1615999
|
0.822
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chr4_-_176970407
|
0.815
|
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_99809068
|
0.814
|
|
PILRA
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr1_+_36121400
|
0.814
|
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr4_-_176970372
|
0.810
|
|
GPM6A
|
glycoprotein M6A
|
|
chr3_+_155321826
|
0.810
|
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr21_-_43903755
|
0.807
|
NM_007031
|
HSF2BP
|
heat shock transcription factor 2 binding protein
|
|
chr5_+_169592523
|
0.806
|
NM_001102609
|
C5orf58
|
chromosome 5 open reading frame 58
|
|
chrX_+_49013424
|
0.806
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chrX_-_152591936
|
0.804
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr11_+_67984758
|
0.803
|
NM_001164160
NM_001164161
NM_001164162
NM_001164163
NM_001164164
NM_018312
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr22_+_20316968
|
0.801
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr7_-_142369534
|
0.800
|
NM_000420
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr11_+_2279932
|
0.797
|
|
TSPAN32
|
tetraspanin 32
|
|
chrX_+_48427103
|
0.796
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr1_+_163780063
|
0.787
|
NM_001005214
|
LRRC52
|
leucine rich repeat containing 52
|
|
chr7_-_142369519
|
0.787
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr16_+_29892714
|
0.786
|
NM_004783
NM_016151
|
TAOK2
|
TAO kinase 2
|
|
chr22_+_18388602
|
0.785
|
NM_152906
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr7_+_73826244
|
0.783
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr11_-_118405288
|
0.781
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr20_+_42417752
|
0.778
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_+_31799099
|
0.775
|
NM_025260
NM_138272
NM_138273
NM_138274
NM_138275
NM_138277
|
C6orf25
|
chromosome 6 open reading frame 25
|
|
chr17_-_9870292
|
0.773
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr8_-_134653194
|
0.769
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr2_+_27225362
|
0.769
|
NM_175769
|
TCF23
|
transcription factor 23
|
|
chr3_-_109292444
|
0.767
|
NM_001777
NM_198793
|
CD47
|
CD47 molecule
|
|
chr11_-_83071084
|
0.767
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr8_+_124264054
|
0.766
|
|
FAM83A
|
family with sequence similarity 83, member A
|
|
chr9_+_135277264
|
0.766
|
|
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13
|
|
chr19_-_60483256
|
0.764
|
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1
|
|
chr16_+_66236559
|
0.758
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr21_+_43903803
|
0.755
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr11_+_19691397
|
0.747
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr19_+_14405100
|
0.745
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr3_+_187917791
|
0.745
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr3_-_39209075
|
0.740
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr2_-_242096673
|
0.740
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr16_+_29581783
|
0.740
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr20_-_43972061
|
0.738
|
|
PLTP
|
phospholipid transfer protein
|
|
chr20_+_34523555
|
0.737
|
NM_183006
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr19_-_12802206
|
0.736
|
NM_001080997
|
RTBDN
|
retbindin
|
|
chr2_+_220087135
|
0.736
|
NM_018674
NM_182847
|
ACCN4
|
amiloride-sensitive cation channel 4, pituitary
|
|
chr2_-_242096592
|
0.735
|
|
STK25
|
serine/threonine kinase 25
|
|
chr15_+_65145215
|
0.731
|
|
SMAD3
|
SMAD family member 3
|
|
chr22_+_18388656
|
0.731
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr3_+_47004457
|
0.730
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chr3_-_185578621
|
0.729
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr15_+_38923914
|
0.727
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr20_-_34102250
|
0.724
|
|
LOC647979
|
hypothetical LOC647979
|
|
chr16_+_66236512
|
0.723
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr19_+_3490154
|
0.720
|
NM_001135580
|
C19orf71
|
chromosome 19 open reading frame 71
|
|
chr6_+_3009120
|
0.715
|
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1
|
|
chrX_+_153062918
|
0.714
|
NM_020061
|
OPN1LW
|
opsin 1 (cone pigments), long-wave-sensitive
|
|
chr1_-_158815895
|
0.701
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr15_-_76313896
|
0.701
|
NM_015162
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr16_-_82778071
|
0.700
|
NM_005679
NM_139353
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa
|
|
chr5_-_149515331
|
0.699
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr7_+_99793561
|
0.699
|
NM_178238
|
PILRB
|
paired immunoglobin-like type 2 receptor beta
|
|
chr7_+_142695584
|
0.696
|
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr1_-_159099090
|
0.691
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr11_+_183079
|
0.690
|
NM_145651
|
SCGB1C1
|
secretoglobin, family 1C, member 1
|
|
chr4_-_176971175
|
0.690
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr16_+_28903647
|
0.687
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr17_-_45617899
|
0.684
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr17_-_35975227
|
0.674
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr10_+_85923473
|
0.673
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr22_-_49042065
|
0.668
|
NM_002969
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr3_-_49701085
|
0.666
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr10_+_85944383
|
0.666
|
NM_001171971
NM_033100
|
CDHR1
|
cadherin-related family member 1
|
|
chr19_-_56260135
|
0.661
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr7_-_92686773
|
0.658
|
NM_198151
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr2_-_242096122
|
0.655
|
|
STK25
|
serine/threonine kinase 25
|
|
chr17_-_59817575
|
0.653
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr7_-_102044374
|
0.652
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr11_+_63030131
|
0.651
|
|
LGALS12
|
lectin, galactoside-binding, soluble, 12
|
|
chr19_-_45611110
|
0.651
|
NM_020956
NM_181882
|
PRX
|
periaxin
|
|
chr12_-_121551424
|
0.651
|
NM_017612
|
ZCCHC8
|
zinc finger, CCHC domain containing 8
|
|
chr1_-_39929654
|
0.650
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr5_-_1647617
|
0.650
|
|
SDHAP3
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3
|
|
chr7_+_94130752
|
0.649
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_35331435
|
0.644
|
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr2_+_128175066
|
0.641
|
NM_032740
|
SFT2D3
|
SFT2 domain containing 3
|
|
chr17_+_35484204
|
0.636
|
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr16_+_28903870
|
0.635
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr8_+_145700272
|
0.634
|
NM_005309
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase)
|
|
chr14_+_95222506
|
0.633
|
NM_004918
NM_199206
|
TCL1B
|
T-cell leukemia/lymphoma 1B
|
|
chr20_+_42417502
|
0.631
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr1_-_202921103
|
0.625
|
NM_006338
NM_201630
|
LRRN2
|
leucine rich repeat neuronal 2
|
|
chr13_-_40138607
|
0.624
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr7_+_43118721
|
0.624
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr1_+_159959072
|
0.623
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr12_+_6517600
|
0.623
|
|
|
|