|
chr19_+_51493486
|
1.298
|
NM_022462
|
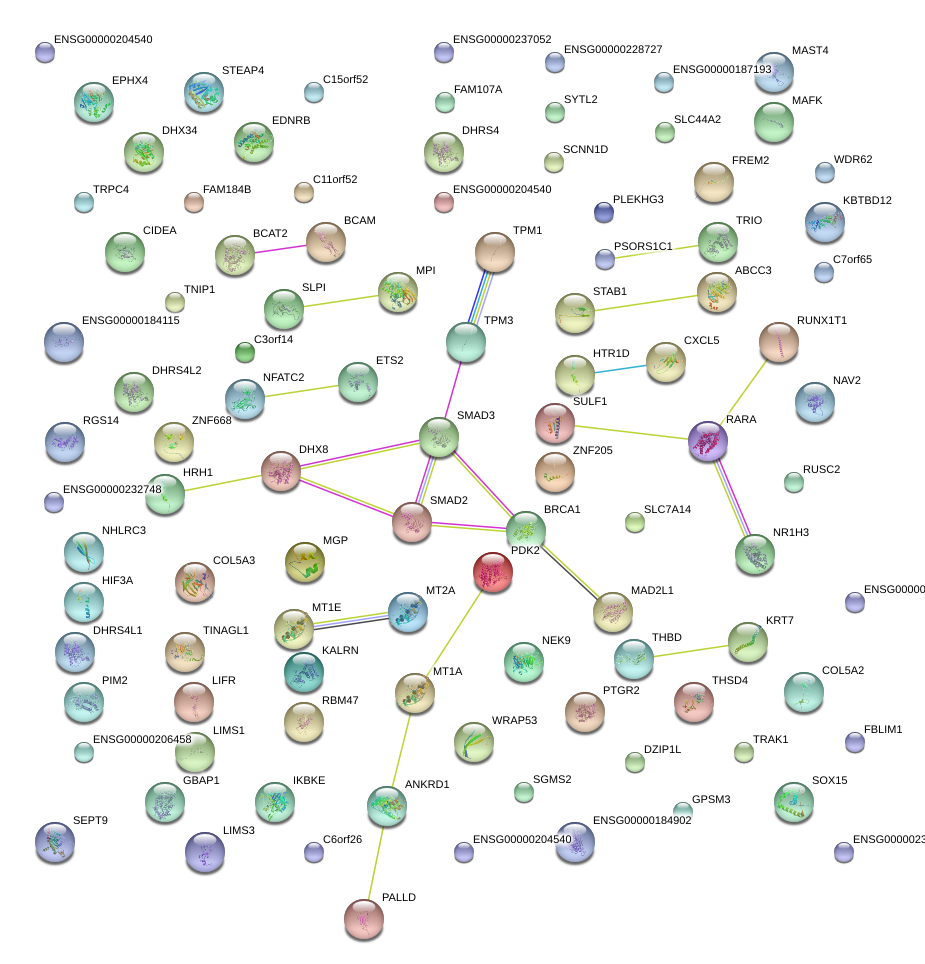
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr1_-_23393808
|
0.761
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr15_+_65205107
|
0.757
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr7_+_1543644
|
0.702
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr12_+_50913274
|
0.702
|
|
KRT7
|
keratin 7
|
|
chr11_+_20001130
|
0.664
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr9_+_35528628
|
0.609
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr3_-_139315011
|
0.607
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr12_+_50913220
|
0.599
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr1_+_31814672
|
0.595
|
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr11_+_20000938
|
0.591
|
|
NAV2
|
neuron navigator 2
|
|
chr1_+_31814709
|
0.589
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr12_-_14929991
|
0.567
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr5_-_38631263
|
0.560
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr5_+_14493905
|
0.518
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr6_+_31190505
|
0.518
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr19_+_50004134
|
0.506
|
NM_001013257
NM_005581
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group)
|
|
chr17_-_7434113
|
0.502
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr9_+_35528890
|
0.486
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr1_+_92268072
|
0.482
|
NM_173567
|
EPHX4
|
epoxide hydrolase 4
|
|
chr15_-_38417564
|
0.478
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr1_+_31814712
|
0.464
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr13_-_37341859
|
0.457
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr17_+_46067114
|
0.452
|
NM_001144070
NM_003786
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr4_-_75083160
|
0.451
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr3_+_11153778
|
0.445
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr8_-_95720870
|
0.443
|
|
LOC100288748
|
hypothetical LOC100288748
|
|
chr13_-_77390907
|
0.442
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr15_+_61123327
|
0.439
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr21_+_39117064
|
0.425
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr5_+_176726918
|
0.419
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr5_-_150441150
|
0.414
|
NM_006058
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr3_+_125586241
|
0.407
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr1_+_15955719
|
0.403
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr11_+_47236072
|
0.398
|
NM_001130101
NM_005693
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr3_+_42176662
|
0.385
|
NM_014965
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr20_-_22978141
|
0.383
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr17_+_35728070
|
0.379
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr19_+_10597167
|
0.376
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr4_+_169789327
|
0.375
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr14_+_23527866
|
0.372
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr16_-_30983909
|
0.371
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr8_+_70567581
|
0.371
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr8_+_70567634
|
0.369
|
|
SULF1
|
sulfatase 1
|
|
chr3_+_129117007
|
0.369
|
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr15_+_69626594
|
0.367
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr15_+_72969404
|
0.362
|
NM_002435
|
MPI
|
mannose phosphate isomerase
|
|
chr11_+_111294868
|
0.361
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr4_-_40326603
|
0.357
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr4_-_121207338
|
0.357
|
NM_002358
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast)
|
|
chr5_+_66160359
|
0.354
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr14_+_64240902
|
0.348
|
NM_015549
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr3_-_58538110
|
0.346
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr10_-_92670764
|
0.345
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr4_+_109033868
|
0.344
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr2_+_110013297
|
0.342
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr1_+_204710453
|
0.332
|
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr17_+_35728039
|
0.332
|
|
RARA
|
retinoic acid receptor, alpha
|
|
chr13_+_38159172
|
0.331
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr3_-_171786468
|
0.329
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr1_-_153463815
|
0.328
|
|
GBAP1
|
glucosidase, beta, acid pseudogene 1
|
|
chr14_+_73388259
|
0.327
|
NM_001146154
NM_001146155
NM_152444
|
PTGR2
|
prostaglandin reductase 2
|
|
chr6_+_31838751
|
0.325
|
NM_001039651
|
C6orf26
|
chromosome 6 open reading frame 26
|
|
chr2_-_189752575
|
0.320
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_+_52504410
|
0.313
|
|
STAB1
|
stabilin 1
|
|
chr7_-_87774130
|
0.313
|
NM_024636
|
STEAP4
|
STEAP family member 4
|
|
chr1_+_204710409
|
0.312
|
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon
|
|
chr6_-_32268267
|
0.311
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr20_-_49612745
|
0.310
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr19_-_9982041
|
0.309
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr7_+_47661366
|
0.309
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr11_-_85108030
|
0.309
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr8_-_93144366
|
0.308
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_+_12244317
|
0.307
|
NM_001279
|
CIDEA
|
cell death-inducing DFFA-like effector a
|
|
chrX_-_48661226
|
0.305
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr17_-_38575737
|
0.301
|
|
BRCA1
|
breast cancer 1, early onset
|
|
chr16_+_55217180
|
0.294
|
|
MT1E
|
metallothionein 1E
|
|
chr14_-_74663376
|
0.294
|
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr1_+_1207351
|
0.293
|
NM_001130413
NM_002978
|
SCNN1D
|
sodium channel, nonvoltage-gated 1, delta
|
|
chr4_-_17392232
|
0.292
|
NM_015688
|
FAM184B
|
family with sequence similarity 184, member B
|
|
chr17_+_45527507
|
0.291
|
NM_002611
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr13_+_38510440
|
0.291
|
NM_001012754
NM_001017370
|
NHLRC3
|
NHL repeat containing 3
|
|
chr16_+_3102556
|
0.286
|
NM_001042428
NM_003456
|
ZNF205
|
zinc finger protein 205
|
|
chr17_+_7532391
|
0.284
|
NM_001143992
NM_018081
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr3_+_62280435
|
0.283
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr15_-_40236061
|
0.281
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr13_-_35327798
|
0.279
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr10_-_92671005
|
0.277
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr2_+_27705018
|
0.277
|
NM_001145047
NM_001145048
NM_001145049
|
GPN1
|
GPN-loop GTPase 1
|
|
chr3_-_17757402
|
0.276
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr13_-_51878235
|
0.275
|
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr17_+_35727998
|
0.274
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr2_-_108972048
|
0.274
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr12_+_121180439
|
0.269
|
|
MLXIP
|
MLX interacting protein
|
|
chr5_-_16795336
|
0.269
|
|
MYO10
|
myosin X
|
|
chr2_-_110665188
|
0.268
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr17_+_7457988
|
0.267
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr14_+_78815406
|
0.265
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr15_-_88256922
|
0.263
|
NM_001199058
NM_182616
|
C15orf38-AP3S2
C15orf38
|
C15orf38-AP3S2 readthrough
chromosome 15 open reading frame 38
|
|
chr12_+_64504408
|
0.263
|
NM_003483
NM_003484
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr19_-_18246166
|
0.263
|
NM_001145304
NM_001145305
NM_025249
|
KIAA1683
|
KIAA1683
|
|
chr21_-_36774257
|
0.260
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chrX_-_74292800
|
0.258
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chr6_-_75972473
|
0.258
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr14_+_64240996
|
0.257
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr13_-_98202884
|
0.255
|
NM_005073
|
SLC15A1
|
solute carrier family 15 (oligopeptide transporter), member 1
|
|
chr15_+_39907447
|
0.254
|
NM_001114632
NM_001198588
NM_005090
|
JMJD7
JMJD7-PLA2G4B
|
jumonji domain containing 7
JMJD7-PLA2G4B readthrough
|
|
chr2_-_128138435
|
0.252
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr16_+_55261226
|
0.249
|
NM_005951
|
MT1H
|
metallothionein 1H
|
|
chr1_-_228916486
|
0.249
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr17_-_39651189
|
0.246
|
NM_014233
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr16_-_11587647
|
0.246
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr11_+_111294810
|
0.243
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr9_-_5620070
|
0.242
|
|
C9orf46
|
chromosome 9 open reading frame 46
|
|
chr1_+_19842309
|
0.242
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr11_+_85017269
|
0.241
|
NM_001193537
NM_001193538
NM_018480
|
TMEM126B
|
transmembrane protein 126B
|
|
chr4_+_128773529
|
0.241
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr12_+_6363482
|
0.240
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr7_+_89621624
|
0.239
|
NM_012449
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1
|
|
chr1_-_228916470
|
0.237
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr10_+_30763056
|
0.236
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr2_+_32142083
|
0.236
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr16_+_1524162
|
0.235
|
NM_024600
|
TMEM204
|
transmembrane protein 204
|
|
chr10_+_104145421
|
0.235
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr10_+_96295511
|
0.234
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr14_-_74663473
|
0.233
|
NM_033116
|
NEK9
|
NIMA (never in mitosis gene a)- related kinase 9
|
|
chr12_+_39372456
|
0.233
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr10_+_52421120
|
0.232
|
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr19_-_49523747
|
0.228
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr20_+_34206070
|
0.228
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr9_+_17568952
|
0.228
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr19_-_49552638
|
0.228
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr3_-_102194938
|
0.227
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr12_-_55013987
|
0.226
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr1_+_3378030
|
0.225
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr6_-_94185780
|
0.225
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr4_+_140442118
|
0.225
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr3_+_144203061
|
0.224
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr12_-_122155998
|
0.223
|
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr11_+_5714253
|
0.222
|
NM_001005180
|
OR56B1
|
olfactory receptor, family 56, subfamily B, member 1
|
|
chr16_+_66129481
|
0.221
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr16_+_1523667
|
0.221
|
|
TMEM204
|
transmembrane protein 204
|
|
chr2_-_171998681
|
0.220
|
|
METTL8
|
methyltransferase like 8
|
|
chr19_-_56147971
|
0.219
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr13_-_110011866
|
0.219
|
NM_017817
|
RAB20
|
RAB20, member RAS oncogene family
|
|
chr5_+_49998677
|
0.219
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr14_+_23575460
|
0.218
|
NM_001082488
|
DHRS4L1
|
dehydrogenase/reductase (SDR family) member 4 like 1
|
|
chr9_+_111927601
|
0.218
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr5_-_125958503
|
0.218
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr5_+_72897323
|
0.217
|
NM_032175
|
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae)
|
|
chr17_-_15106597
|
0.217
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr11_-_10672079
|
0.216
|
NM_001100163
NM_130385
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr15_-_66511488
|
0.216
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr17_+_63144500
|
0.216
|
NM_015462
|
NOL11
|
nucleolar protein 11
|
|
chr3_-_69518119
|
0.214
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr17_-_74433048
|
0.214
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr17_-_15106579
|
0.214
|
|
PMP22
|
peripheral myelin protein 22
|
|
chr11_-_57792206
|
0.213
|
NM_207374
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1
|
|
chr2_+_108637698
|
0.212
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr19_-_46626474
|
0.212
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr4_-_5881271
|
0.210
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr16_+_55217085
|
0.210
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr2_-_105312536
|
0.209
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr14_+_95740916
|
0.207
|
|
BDKRB2
|
bradykinin receptor B2
|
|
chr16_-_87244896
|
0.206
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr8_-_81246215
|
0.205
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr4_-_153675602
|
0.205
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr21_-_46529663
|
0.203
|
NM_003906
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr10_-_120914825
|
0.203
|
|
SFXN4
|
sideroflexin 4
|
|
chr10_+_53744043
|
0.202
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr9_+_4975082
|
0.202
|
|
JAK2
|
Janus kinase 2
|
|
chr1_-_228916559
|
0.201
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_+_144227916
|
0.201
|
NM_001184795
|
PEX11B
|
peroxisomal biogenesis factor 11 beta
|
|
chr19_+_60691671
|
0.200
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr16_-_69277194
|
0.200
|
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chrX_+_73440749
|
0.199
|
NM_203303
|
ZCCHC13
|
zinc finger, CCHC domain containing 13
|
|
chr14_-_76958984
|
0.198
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr1_-_6473216
|
0.198
|
NM_001042664
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr2_-_238164042
|
0.197
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr22_-_30018329
|
0.196
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr9_+_8848130
|
0.195
|
|
|
|
|
chr18_+_18767764
|
0.195
|
NM_203291
NM_203292
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr1_-_32940943
|
0.194
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr16_-_66902286
|
0.194
|
NM_032178
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand
|
|
chr9_+_35739276
|
0.193
|
NM_001080496
|
RGP1
|
RGP1 retrograde golgi transport homolog (S. cerevisiae)
|
|
chr3_-_12561962
|
0.193
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr2_+_108637916
|
0.192
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr7_+_75977680
|
0.192
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr17_+_38575939
|
0.191
|
NM_031862
NM_031858
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr15_-_70838703
|
0.191
|
|
|
|
|
chr3_-_193609531
|
0.191
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr19_-_16631905
|
0.187
|
|
C19orf42
|
chromosome 19 open reading frame 42
|
|
chr11_+_124967961
|
0.187
|
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr16_+_56219802
|
0.187
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr12_-_111114908
|
0.187
|
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr4_-_140443061
|
0.187
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr19_+_41419165
|
0.186
|
|
ZNF146
|
zinc finger protein 146
|
|
chr11_+_124967946
|
0.186
|
NM_152713
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr8_+_79591074
|
0.185
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr20_-_23808313
|
0.185
|
NM_001900
|
CST5
|
cystatin D
|