|
chrX_+_135079120
|
2.477
|
NM_001159704
|
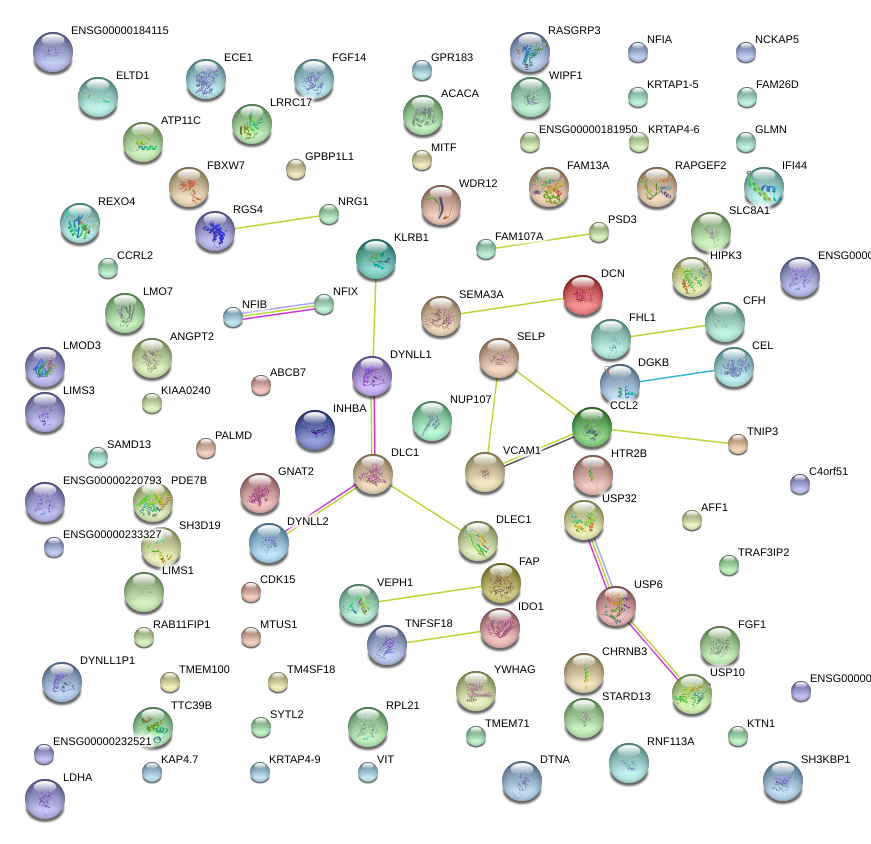
FHL1
|
four and a half LIM domains 1
|
|
chr1_+_84539876
|
1.989
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_+_69895310
|
1.836
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_15240113
|
1.695
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr6_+_42896771
|
1.539
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr8_-_13416554
|
1.533
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr17_+_36515166
|
1.514
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr1_+_78888064
|
1.451
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr4_-_153523113
|
1.439
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_-_79244948
|
1.420
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr1_+_161305558
|
1.409
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr7_+_102340579
|
1.390
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr3_-_158695832
|
1.363
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_-_167865973
|
1.348
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr4_-_122357104
|
1.321
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr6_+_116956868
|
1.321
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr8_+_42671718
|
1.291
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr8_-_6407882
|
1.285
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr1_-_109957195
|
1.274
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr7_-_75794919
|
1.273
|
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr8_-_133841988
|
1.236
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr13_-_101852028
|
1.226
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr3_-_150533948
|
1.221
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr1_+_61642287
|
1.202
|
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_51155073
|
1.201
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr1_+_99884018
|
1.193
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr2_-_40593074
|
1.186
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_+_202379442
|
1.184
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr4_+_88187215
|
1.179
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr17_-_32730804
|
1.176
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr2_-_134042500
|
1.164
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr18_+_30544193
|
1.142
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr1_+_19807168
|
1.125
|
|
|
|
|
chr2_-_162808123
|
1.122
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr3_+_69998064
|
1.122
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr2_+_33514896
|
1.120
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr13_-_98757634
|
1.095
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr8_-_37847095
|
1.093
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr2_-_175170674
|
1.088
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr8_+_32698892
|
1.073
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_+_129682434
|
1.062
|
|
|
|
|
chrX_-_138742050
|
1.015
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr8_+_39890484
|
1.014
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr3_+_46423682
|
1.002
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr3_+_69895651
|
0.969
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_-_132300702
|
0.968
|
|
|
|
|
chr10_-_112668679
|
0.958
|
NM_001195306
|
BBIP1
|
BBSome interacting protein 1
|
|
chr17_+_29606408
|
0.953
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr5_-_142057692
|
0.944
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_-_231697999
|
0.941
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr1_+_22107137
|
0.939
|
|
RPL21
|
ribosomal protein L21
|
|
chr1_+_100957883
|
0.937
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr6_+_136214495
|
0.931
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_203484445
|
0.927
|
|
WDR12
|
WD repeat domain 12
|
|
chr4_-_89963177
|
0.917
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr1_-_45868775
|
0.917
|
|
GPBP1L1
|
GC-rich promoter binding protein 1-like 1
|
|
chr2_+_108571336
|
0.915
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr6_-_111995213
|
0.899
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr11_-_85115105
|
0.886
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr12_-_90097384
|
0.881
|
NM_133503
|
DCN
|
decorin
|
|
chr13_+_75092570
|
0.880
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr3_-_69254226
|
0.879
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr17_+_36493927
|
0.871
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr1_-_21478571
|
0.869
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr1_-_171286634
|
0.866
|
NM_005092
|
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18
|
|
chr1_+_161305188
|
0.845
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr4_+_146820805
|
0.839
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr12_+_67366989
|
0.839
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr2_+_36777406
|
0.834
|
|
VIT
|
vitrin
|
|
chr7_-_83662152
|
0.830
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr2_+_108571177
|
0.822
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_-_18585721
|
0.819
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr11_+_18374388
|
0.817
|
NM_001165414
|
LDHA
|
lactate dehydrogenase A
|
|
chr11_+_33306652
|
0.816
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chrX_-_74292800
|
0.815
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chr7_-_14847479
|
0.814
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr13_-_32678142
|
0.814
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_36436979
|
0.810
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr16_+_83349816
|
0.807
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chrX_-_19598912
|
0.805
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_+_160408447
|
0.799
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr3_-_58538110
|
0.799
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr8_-_17599438
|
0.785
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr14_+_55148488
|
0.784
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chrX_+_135079461
|
0.780
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_-_152366970
|
0.774
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr7_-_41709191
|
0.773
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr5_-_34078991
|
0.771
|
NM_030945
NM_181435
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3
|
|
chr4_-_24590867
|
0.761
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr17_-_23244499
|
0.758
|
NM_001076680
|
C17orf108
|
chromosome 17 open reading frame 108
|
|
chr12_+_91654395
|
0.757
|
NM_001004330
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7
|
|
chr6_-_32447626
|
0.756
|
NM_006781
|
C6orf10
|
chromosome 6 open reading frame 10
|
|
chr10_+_69539255
|
0.756
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr4_-_101658094
|
0.755
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr1_+_169326641
|
0.750
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr5_+_145296313
|
0.747
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr4_+_95595419
|
0.747
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr6_+_106653429
|
0.744
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chrX_+_149364377
|
0.738
|
NM_001177466
NM_005491
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr2_+_108571209
|
0.732
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr18_+_54013575
|
0.729
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr1_-_109201228
|
0.725
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr15_-_40537002
|
0.725
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr4_+_14985272
|
0.724
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr20_-_17487480
|
0.722
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr2_-_202271598
|
0.719
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr10_+_95838801
|
0.719
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr11_+_5597749
|
0.711
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr17_-_64649608
|
0.708
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr13_+_96592051
|
0.702
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_+_157246305
|
0.697
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr14_-_49932366
|
0.694
|
NM_004196
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase)
|
|
chr1_-_23393808
|
0.693
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr11_-_58099884
|
0.690
|
NM_004811
|
LPXN
|
leupaxin
|
|
chr12_-_15706340
|
0.690
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_-_112186977
|
0.683
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr6_+_129245978
|
0.677
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr15_+_60838053
|
0.677
|
|
TLN2
|
talin 2
|
|
chr2_-_144994349
|
0.677
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_+_154120170
|
0.676
|
|
SYT11
|
synaptotagmin XI
|
|
chr1_-_149047329
|
0.674
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr10_+_63092724
|
0.673
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr1_-_155281785
|
0.669
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chr7_-_110989582
|
0.666
|
NM_032549
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr17_-_64462976
|
0.660
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr9_-_90983195
|
0.660
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr9_+_112470871
|
0.657
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr7_+_93388951
|
0.655
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr8_-_125648576
|
0.654
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr11_-_102174032
|
0.654
|
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr9_+_129893948
|
0.652
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr7_-_27133163
|
0.650
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr21_-_38210531
|
0.650
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr9_+_676640
|
0.649
|
|
|
|
|
chr5_+_150008030
|
0.643
|
|
SYNPO
|
synaptopodin
|
|
chr2_-_40510923
|
0.642
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr9_-_94284455
|
0.639
|
|
ASPN
|
asporin
|
|
chr15_-_70350609
|
0.639
|
NM_020214
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6
|
|
chr2_+_102320148
|
0.639
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr3_-_188935363
|
0.629
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr12_-_52281050
|
0.620
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr11_-_114593838
|
0.617
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr22_-_37211467
|
0.615
|
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr1_+_17817397
|
0.611
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr1_+_161308318
|
0.611
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr10_+_104525877
|
0.609
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr2_-_99412694
|
0.607
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr18_-_51219880
|
0.605
|
|
TCF4
|
transcription factor 4
|
|
chr5_+_147775652
|
0.605
|
|
FBXO38
|
F-box protein 38
|
|
chr5_+_152850276
|
0.604
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr9_+_35528890
|
0.603
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr7_+_134226690
|
0.598
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr1_-_194844061
|
0.598
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr17_-_36451189
|
0.597
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr1_+_194887630
|
0.595
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr10_-_35419229
|
0.585
|
NM_003591
|
CUL2
|
cullin 2
|
|
chr12_+_12829807
|
0.583
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr8_-_103735194
|
0.583
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr11_-_102174102
|
0.582
|
NM_001145938
NM_002421
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase)
|
|
chr12_+_118100977
|
0.582
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr5_+_76470829
|
0.580
|
|
|
|
|
chr15_-_68781662
|
0.577
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr19_-_47961653
|
0.577
|
NM_001130167
NM_001130168
NM_182707
|
PSG8
|
pregnancy specific beta-1-glycoprotein 8
|
|
chr13_+_51334117
|
0.577
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr21_-_38792173
|
0.575
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr21_-_34749908
|
0.574
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr2_-_101134146
|
0.573
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr3_-_188936941
|
0.569
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_213723298
|
0.568
|
NM_016260
|
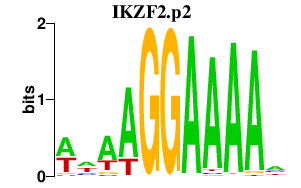
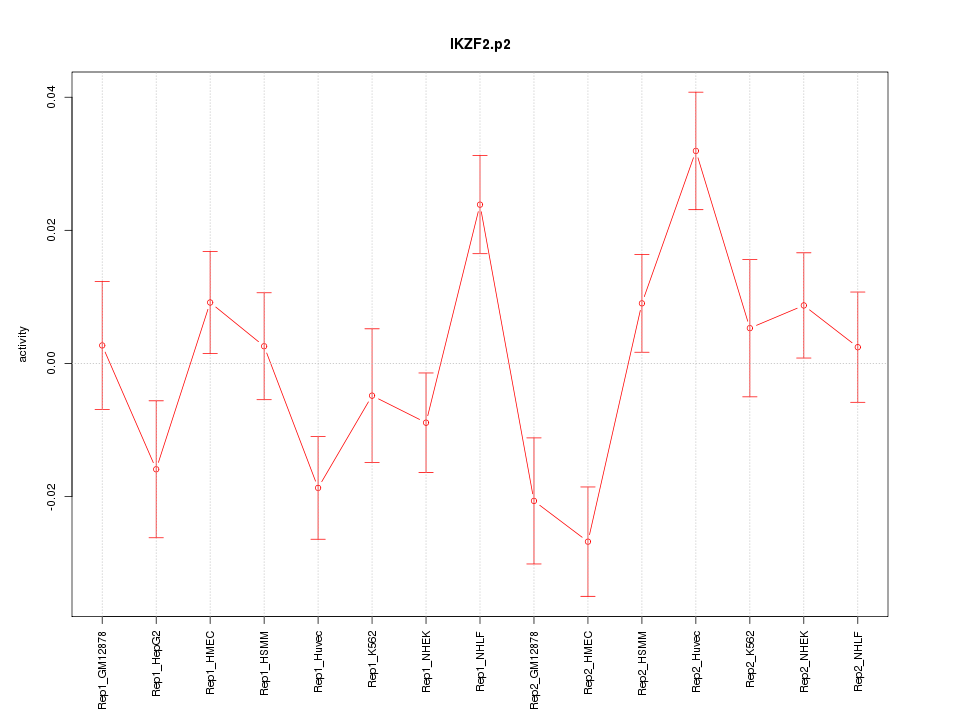
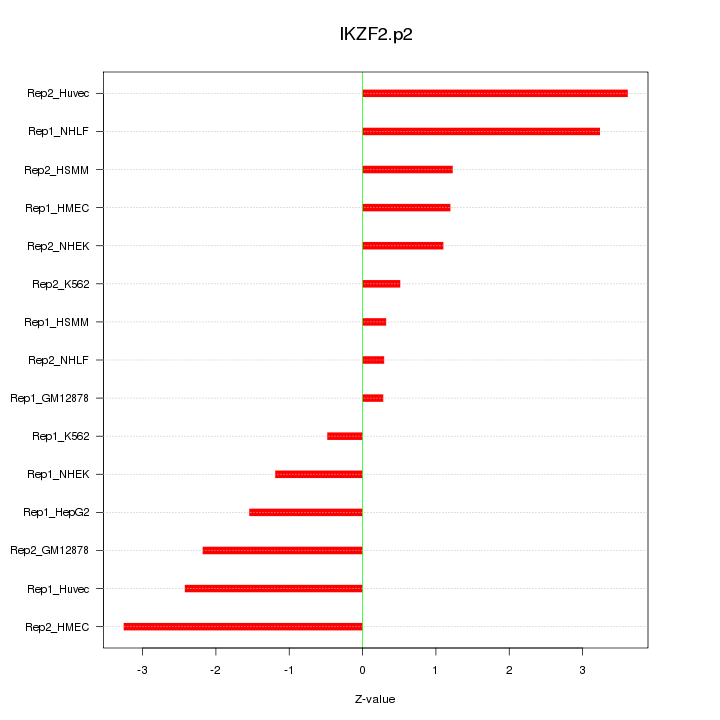
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr6_-_56615544
|
0.567
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr13_-_37070862
|
0.566
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr3_-_11585264
|
0.565
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr14_+_38720085
|
0.563
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr5_-_58607673
|
0.563
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_14929991
|
0.560
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr8_-_82606104
|
0.560
|
NM_001105281
|
FABP12
|
fatty acid binding protein 12
|
|
chr1_+_145841569
|
0.559
|
NM_005267
|
GJA8
|
gap junction protein, alpha 8, 50kDa
|
|
chr14_+_85066265
|
0.557
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr12_-_50871953
|
0.556
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr13_-_40735619
|
0.555
|
NM_004294
|
MTRF1
|
mitochondrial translational release factor 1
|
|
chr12_-_78853365
|
0.554
|
NM_001143885
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chrX_+_56772391
|
0.553
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr20_+_56369012
|
0.552
|
|
|
|
|
chr1_-_151804929
|
0.552
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr1_+_93317374
|
0.551
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chrX_-_135166306
|
0.550
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr21_-_35174742
|
0.547
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr4_+_15080586
|
0.546
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr1_+_159435728
|
0.545
|
NM_004550
|
NDUFS2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
|
|
chr16_+_31105579
|
0.545
|
|
|
|
|
chr2_-_189363075
|
0.545
|
NM_052952
|
DIRC1
|
disrupted in renal carcinoma 1
|
|
chr10_-_29963906
|
0.542
|
NM_021738
|
SVIL
|
supervillin
|
|
chr12_-_7487989
|
0.542
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr7_+_90177082
|
0.538
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr7_-_137271191
|
0.536
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr5_-_59100194
|
0.532
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_+_91762227
|
0.531
|
|
|
|
|
chr10_+_102285690
|
0.531
|
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr9_+_35528628
|
0.530
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr10_-_116434364
|
0.530
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|