|
chr10_+_80672842
|
5.493
|
|
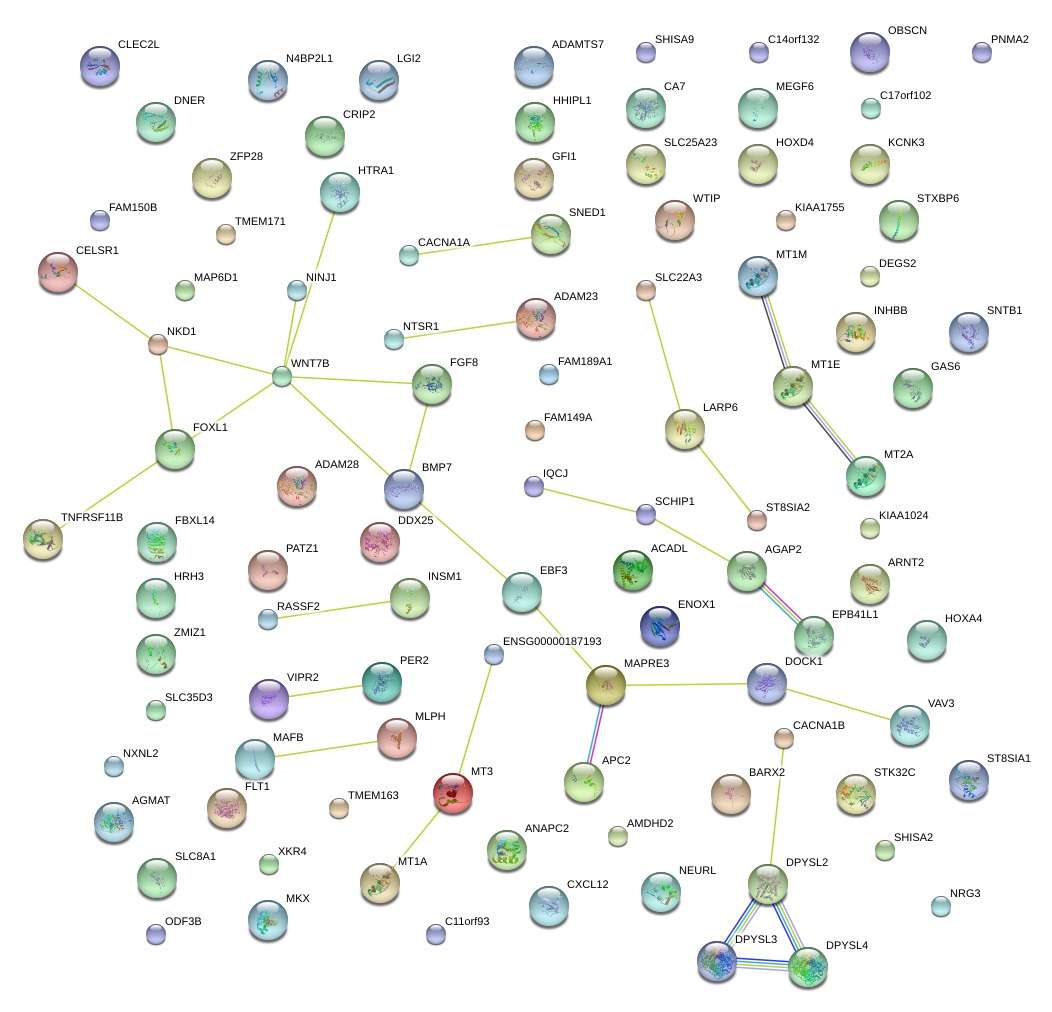
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr15_-_27650218
|
4.412
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr2_+_238060602
|
4.331
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr9_+_90339835
|
3.821
|
NM_001161625
NM_145283
|
NXNL2
|
nucleoredoxin-like 2
|
|
chr8_-_121893468
|
3.778
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr16_+_2510323
|
3.707
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr22_-_49317408
|
3.571
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr14_+_99181232
|
3.499
|
NM_001127258
NM_032425
|
HHIPL1
|
HHIP-like 1
|
|
chr10_-_128884411
|
3.439
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr3_-_185025989
|
3.313
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr15_+_77511912
|
3.291
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr2_+_120820140
|
3.203
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr10_+_124210980
|
3.179
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr16_+_55224034
|
3.136
|
NM_176870
|
MT1M
|
metallothionein 1M
|
|
chr12_-_56418295
|
3.105
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr20_+_34206070
|
2.874
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr20_-_4752067
|
2.848
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr12_-_1573591
|
2.754
|
NM_152441
|
FBXL14
|
F-box and leucine-rich repeat protein 14
|
|
chr16_+_55180767
|
2.741
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr15_-_68933481
|
2.730
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chr10_-_135088065
|
2.674
|
NM_001012508
|
SPRN
|
shadow of prion protein homolog (zebrafish)
|
|
chr8_-_121893449
|
2.596
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr16_+_85169615
|
2.594
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr2_+_26768894
|
2.587
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr20_-_36322587
|
2.582
|
NM_001029864
|
KIAA1755
|
KIAA1755
|
|
chr20_+_20296744
|
2.569
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr16_+_55217180
|
2.550
|
|
MT1E
|
metallothionein 1E
|
|
chr10_+_133850325
|
2.546
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr2_-_40532651
|
2.534
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_-_238861830
|
2.530
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr4_+_187302988
|
2.508
|
NM_015398
|
FAM149A
|
family with sequence similarity 149, member A
|
|
chr7_-_27136876
|
2.483
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr16_+_49139694
|
2.472
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr10_-_133971136
|
2.436
|
NM_173575
|
STK32C
|
serine/threonine kinase 32C
|
|
chr17_-_5344898
|
2.399
|
NM_001162371
|
LOC728392
|
hypothetical protein LOC728392
|
|
chr8_-_120033240
|
2.387
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr11_+_110675175
|
2.379
|
NM_001136105
|
C11orf93
|
chromosome 11 open reading frame 93
|
|
chr8_+_56177567
|
2.370
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr12_-_22378833
|
2.367
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr13_-_43259032
|
2.340
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr13_+_113587674
|
2.334
|
|
GAS6
|
growth arrest-specific 6
|
|
chr20_-_60228665
|
2.326
|
NM_007232
|
HRH3
|
histamine receptor H3
|
|
chr14_+_105012106
|
2.286
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr2_-_230287442
|
2.274
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr10_+_105243724
|
2.270
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr13_-_25523164
|
2.269
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr10_-_44200458
|
2.267
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr2_-_210798232
|
2.247
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr16_+_55217085
|
2.204
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr10_-_28074752
|
2.196
|
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr12_+_56406296
|
2.192
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr15_+_78483624
|
2.189
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr2_+_241586927
|
2.169
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr10_-_103525646
|
2.167
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr15_+_90738108
|
2.156
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr20_-_55274497
|
2.146
|
|
BMP7
|
bone morphogenetic protein 7
|
|
chr13_-_27967215
|
2.129
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr10_+_83625049
|
2.123
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr20_-_38751289
|
2.118
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr6_+_160689355
|
2.108
|
NM_021977
|
SLC22A3
|
solute carrier family 22 (extraneuronal monoamine transporter), member 3
|
|
chr22_-_44751671
|
2.077
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr22_-_30072329
|
2.074
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr16_+_12902955
|
2.066
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr1_-_92721633
|
2.058
|
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr17_-_29930431
|
2.056
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chr1_-_3437834
|
2.020
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr14_+_95575319
|
2.000
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr2_-_135193011
|
1.993
|
NM_030923
|
TMEM163
|
transmembrane protein 163
|
|
chr2_-_39040981
|
1.988
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr19_+_39664706
|
1.977
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr7_+_138859213
|
1.975
|
NM_001080511
|
CLEC2L
|
C-type lectin domain family 2, member L
|
|
chr4_-_24641404
|
1.973
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr22_-_45311730
|
1.972
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr9_-_94936319
|
1.960
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr5_-_146869811
|
1.939
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr10_+_124211353
|
1.938
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_+_160964574
|
1.931
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_-_278181
|
1.926
|
NM_001002919
|
FAM150B
|
family with sequence similarity 150, member B
|
|
chr15_-_76890626
|
1.916
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr22_-_30072217
|
1.916
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr7_-_158630223
|
1.900
|
NM_003382
|
VIPR2
|
vasoactive intestinal peptide receptor 2
|
|
chr8_+_26427378
|
1.898
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr1_-_108308997
|
1.892
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr19_+_1397268
|
1.890
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr8_-_121893196
|
1.888
|
|
|
|
|
chr3_-_62835601
|
1.878
|
|
|
|
|
chr20_+_60810610
|
1.871
|
NM_002531
|
NTSR1
|
neurotensin receptor 1 (high affinity)
|
|
chr14_-_24588916
|
1.866
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr9_+_139892049
|
1.848
|
NM_000718
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr11_+_128751044
|
1.823
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr1_-_15784048
|
1.803
|
NM_024758
|
AGMAT
|
agmatine ureohydrolase (agmatinase)
|
|
chr5_+_72452131
|
1.801
|
NM_001161342
NM_173490
|
TMEM171
|
transmembrane protein 171
|
|
chr10_+_128583984
|
1.795
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr8_-_26427304
|
1.790
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr10_-_131652364
|
1.785
|
|
EBF3
|
early B-cell factor 3
|
|
chr14_-_99695684
|
1.779
|
NM_206918
|
DEGS2
|
degenerative spermatocyte homolog 2, lipid desaturase (Drosophila)
|
|
chr19_+_61742111
|
1.776
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr13_-_31900218
|
1.764
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr6_+_137285071
|
1.762
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chr11_+_125279481
|
1.757
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr8_-_143805371
|
1.753
|
|
LOC100288181
|
hypothetical LOC100288181
|
|
chr16_+_65436341
|
1.744
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr2_+_207016525
|
1.743
|
NM_003812
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr5_+_132111034
|
1.730
|
NM_001039780
|
CCNI2
|
cyclin I family, member 2
|
|
chr12_+_5411534
|
1.721
|
NM_001102654
|
NTF3
|
neurotrophin 3
|
|
chr21_+_44560779
|
1.718
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr1_-_57661373
|
1.709
|
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr14_-_103248193
|
1.708
|
|
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3
|
|
chr13_-_19665100
|
1.701
|
NM_004004
|
GJB2
|
gap junction protein, beta 2, 26kDa
|
|
chr14_+_73073570
|
1.699
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr13_+_43845977
|
1.698
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr5_-_16670085
|
1.695
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr2_-_233501069
|
1.695
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chrX_+_68641802
|
1.687
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr1_+_208472817
|
1.683
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr1_-_23623713
|
1.678
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr7_-_76883642
|
1.675
|
NM_017439
|
PION
|
pigeon homolog (Drosophila)
|
|
chr5_-_146869613
|
1.674
|
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr7_-_27206249
|
1.671
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr18_+_58341637
|
1.669
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr1_+_235272324
|
1.656
|
NM_001035
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr16_+_76379901
|
1.650
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chr6_-_170442090
|
1.642
|
|
|
|
|
chr16_-_55259405
|
1.638
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr2_+_176702667
|
1.637
|
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr19_+_1226510
|
1.627
|
NM_017914
|
C19orf24
|
chromosome 19 open reading frame 24
|
|
chr3_+_71885890
|
1.627
|
NM_018971
|
GPR27
|
G protein-coupled receptor 27
|
|
chr20_-_39428911
|
1.623
|
NM_052846
|
EMILIN3
|
elastin microfibril interfacer 3
|
|
chr17_+_78630793
|
1.605
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr8_+_145461356
|
1.597
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr20_+_13150417
|
1.596
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr16_+_55249355
|
1.594
|
NM_005949
|
MT1F
|
metallothionein 1F
|
|
chr10_-_725522
|
1.591
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr2_+_219532590
|
1.586
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr1_-_38285033
|
1.586
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr22_-_30071565
|
1.585
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr9_-_111300350
|
1.585
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr12_+_2032649
|
1.585
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr16_+_2023279
|
1.571
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr20_-_61639104
|
1.566
|
NM_005975
|
PTK6
|
PTK6 protein tyrosine kinase 6
|
|
chr15_-_23659329
|
1.565
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr11_+_124240491
|
1.556
|
NM_022370
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila)
|
|
chr21_+_41719847
|
1.554
|
NM_002462
NM_001178046
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
|
|
chr2_+_242322645
|
1.552
|
NM_152783
|
D2HGDH
|
D-2-hydroxyglutarate dehydrogenase
|
|
chr18_-_72973698
|
1.550
|
|
MBP
|
myelin basic protein
|
|
chr19_+_951295
|
1.549
|
NM_138690
|
GRIN3B
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B
|
|
chr16_-_49742651
|
1.539
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr8_-_143692814
|
1.529
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr16_+_19087035
|
1.525
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr4_+_54660954
|
1.523
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr2_-_106869488
|
1.520
|
|
|
|
|
chr22_-_23318829
|
1.515
|
NM_207644
|
C22orf36
|
chromosome 22 open reading frame 36
|
|
chr7_-_81910956
|
1.515
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr19_+_38804694
|
1.512
|
NM_001127895
NM_001127896
|
CHST8
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8
|
|
chr7_+_140420500
|
1.512
|
NM_001195278
|
LOC100131199
|
transmembrane protein 178-like
|
|
chrX_-_132376660
|
1.509
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr12_-_119291276
|
1.508
|
NM_002442
|
MSI1
|
musashi homolog 1 (Drosophila)
|
|
chr16_-_87535106
|
1.501
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr6_-_153494076
|
1.500
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr10_+_119292699
|
1.498
|
|
|
|
|
chr11_-_9069668
|
1.495
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr8_-_75396016
|
1.494
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr6_+_35290160
|
1.491
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr17_+_27617307
|
1.486
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr10_+_129595349
|
1.483
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr8_+_145393450
|
1.478
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr19_-_60573552
|
1.477
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr18_+_42168180
|
1.461
|
NM_152470
|
C18orf23
RNF165
|
chromosome 18 open reading frame 23
ring finger protein 165
|
|
chr18_+_74841262
|
1.459
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr14_+_69415880
|
1.456
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr2_+_85834111
|
1.444
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr2_-_232499202
|
1.441
|
NM_024409
|
NPPC
|
natriuretic peptide C
|
|
chr11_-_1725662
|
1.437
|
|
LOC402778
|
CD225 family protein FLJ76511
|
|
chr13_+_110565587
|
1.433
|
NM_001113511
NM_001113512
NM_145735
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr4_+_42094612
|
1.430
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr10_+_71232281
|
1.420
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr1_+_199108705
|
1.416
|
NM_005298
|
GPR25
|
G protein-coupled receptor 25
|
|
chr3_-_48607596
|
1.415
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr8_-_6680429
|
1.412
|
NM_207411
|
XKR5
|
XK, Kell blood group complex subunit-related family, member 5
|
|
chr15_+_87147667
|
1.409
|
NM_001135
NM_013227
|
ACAN
|
aggrecan
|
|
chr20_-_3166834
|
1.404
|
NM_001174090
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr19_-_51081147
|
1.402
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr4_-_46690197
|
1.400
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr12_+_55896844
|
1.393
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr11_+_8059267
|
1.392
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chrX_-_71442488
|
1.375
|
NM_001144885
NM_001144887
|
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1
|
|
chr4_-_122213018
|
1.373
|
NM_024574
|
C4orf31
|
chromosome 4 open reading frame 31
|
|
chr7_-_150495799
|
1.372
|
NM_001098834
|
GBX1
|
gastrulation brain homeobox 1
|
|
chr2_+_25118417
|
1.370
|
NM_014971
|
EFR3B
|
EFR3 homolog B (S. cerevisiae)
|
|
chr7_+_3307445
|
1.368
|
NM_152744
|
SDK1
|
sidekick homolog 1, cell adhesion molecule (chicken)
|
|
chr18_+_42167904
|
1.366
|
|
RNF165
|
ring finger protein 165
|
|
chr17_-_45901173
|
1.365
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr2_+_74595073
|
1.359
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr10_-_88116181
|
1.351
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr17_+_40192072
|
1.343
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr15_-_49174107
|
1.336
|
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr7_-_35260213
|
1.335
|
NM_001077653
NM_001166220
|
TBX20
|
T-box 20
|
|
chr15_-_63457378
|
1.331
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr2_+_176761552
|
1.331
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr9_+_90796169
|
1.326
|
NM_005226
|
S1PR3
|
sphingosine-1-phosphate receptor 3
|