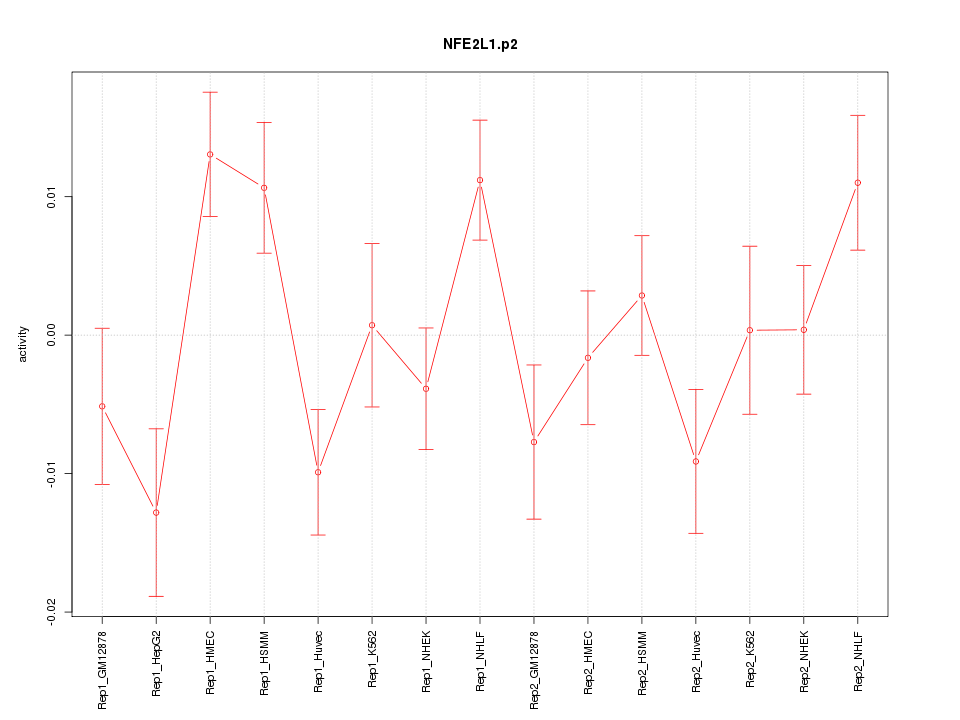
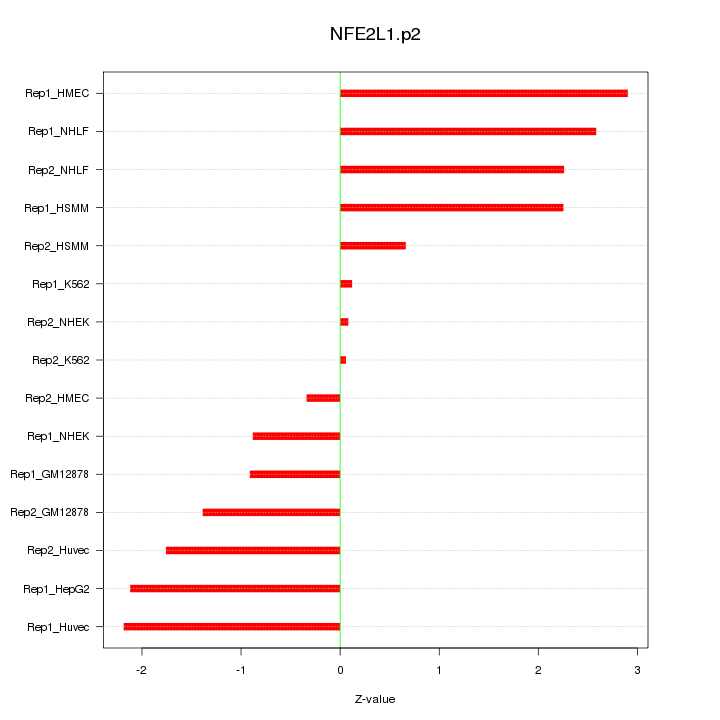
|
chr19_-_40711034
|
2.505
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr4_-_23500706
|
2.439
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr17_-_36475652
|
2.078
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr12_-_88573949
|
1.676
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr5_+_140848905
|
1.558
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr1_+_26973804
|
1.558
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr2_+_220017617
|
1.540
|
|
SPEG
|
SPEG complex locus
|
|
chrX_-_10495476
|
1.463
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_+_201182187
|
1.448
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr5_+_36644187
|
1.410
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr7_+_150442652
|
1.335
|
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3
|
|
chr9_+_35032222
|
1.328
|
NM_001040410
NM_203299
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr17_-_36469869
|
1.303
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr11_-_62213577
|
1.267
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr7_+_129634935
|
1.258
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr8_-_37847095
|
1.232
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr9_-_131555111
|
1.164
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr17_-_51155073
|
1.157
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr11_+_125781157
|
1.156
|
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4
|
|
chr3_-_158695832
|
1.139
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr4_-_187764213
|
1.139
|
|
|
|
|
chr17_-_36877002
|
1.114
|
NM_002278
|
KRT32
|
keratin 32
|
|
chr1_-_215329569
|
1.097
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr7_+_129720209
|
1.096
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr10_+_75343352
|
1.092
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr2_-_74455161
|
1.083
|
NM_001135041
NM_023019
|
DCTN1
|
dynactin 1
|
|
chr15_+_60726801
|
1.082
|
NM_015059
|
TLN2
|
talin 2
|
|
chr5_-_34020445
|
1.078
|
NM_001012509
NM_016180
|
SLC45A2
|
solute carrier family 45, member 2
|
|
chr11_-_85108030
|
1.076
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_16561936
|
1.040
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr7_-_13992351
|
1.037
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr5_-_54317102
|
1.034
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr7_-_135083906
|
1.033
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr8_-_93099040
|
1.011
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_173794692
|
1.009
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr19_+_52113772
|
0.979
|
NM_004491
|
GRLF1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr19_+_10642648
|
0.969
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr11_-_62213946
|
0.942
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr12_-_50871953
|
0.924
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr8_-_72431274
|
0.921
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_-_46401363
|
0.920
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_+_86659370
|
0.907
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr12_+_7038240
|
0.887
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr5_+_166644420
|
0.886
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr8_-_108579270
|
0.886
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr20_+_2743613
|
0.883
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr12_-_51173238
|
0.875
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr5_-_56814310
|
0.851
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr6_-_130224108
|
0.850
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr16_+_66476281
|
0.843
|
NM_198443
|
NRN1L
|
neuritin 1-like
|
|
chr11_-_85107569
|
0.843
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr15_+_40427592
|
0.842
|
NM_212465
|
CAPN3
|
calpain 3, (p94)
|
|
chr3_-_145049853
|
0.841
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr11_+_7574992
|
0.838
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr1_+_154520693
|
0.814
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr15_+_65205107
|
0.811
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chrX_+_105856531
|
0.811
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr3_+_69895310
|
0.810
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr14_+_99555471
|
0.809
|
|
EVL
|
Enah/Vasp-like
|
|
chr6_+_158990982
|
0.808
|
NM_001009991
|
SYTL3
|
synaptotagmin-like 3
|
|
chr11_+_58146721
|
0.805
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr1_+_205137410
|
0.804
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr16_+_56219802
|
0.798
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr12_+_79625538
|
0.796
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr17_+_76549658
|
0.791
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr17_-_7434113
|
0.780
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr4_-_177427252
|
0.780
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr8_+_145718702
|
0.778
|
|
|
|
|
chr17_-_26672757
|
0.775
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr1_-_166965034
|
0.770
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr2_+_220033777
|
0.767
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chrX_-_33267646
|
0.767
|
NM_000109
|
DMD
|
dystrophin
|
|
chr8_+_32698892
|
0.758
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr13_+_23042722
|
0.755
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr19_+_51804260
|
0.753
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr12_-_10215942
|
0.750
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr3_+_63928459
|
0.739
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr8_-_124734346
|
0.737
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr17_-_36996611
|
0.729
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr11_-_123353607
|
0.728
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr22_-_49062461
|
0.726
|
|
PLXNB2
|
plexin B2
|
|
chr17_-_45619038
|
0.713
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr1_-_20682541
|
0.712
|
|
|
|
|
chr5_-_138238877
|
0.705
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr11_-_123772456
|
0.701
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr11_+_5714253
|
0.700
|
NM_001005180
|
OR56B1
|
olfactory receptor, family 56, subfamily B, member 1
|
|
chr2_-_227952249
|
0.696
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr21_-_30510113
|
0.688
|
NM_199328
|
CLDN8
|
claudin 8
|
|
chr5_-_32424444
|
0.687
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr11_-_46674233
|
0.685
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr1_-_23393808
|
0.680
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr4_-_139382672
|
0.677
|
NM_014331
|
SLC7A11
|
solute carrier family 7, (cationic amino acid transporter, y+ system) member 11
|
|
chr9_+_88953378
|
0.674
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr8_-_145090892
|
0.674
|
NM_201381
|
PLEC
|
plectin
|
|
chr11_-_7941617
|
0.669
|
NM_176821
|
NLRP10
|
NLR family, pyrin domain containing 10
|
|
chr6_-_46401174
|
0.668
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr11_-_5523328
|
0.662
|
NM_001005289
|
OR52H1
|
olfactory receptor, family 52, subfamily H, member 1
|
|
chr2_+_88148413
|
0.660
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr5_+_132237253
|
0.659
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr5_+_53787187
|
0.657
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr1_+_159943385
|
0.655
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr15_+_60838053
|
0.651
|
|
TLN2
|
talin 2
|
|
chr1_+_15128882
|
0.651
|
NM_001018001
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr3_+_63613383
|
0.641
|
NM_001080537
|
SNTN
|
sentan, cilia apical structure protein
|
|
chr5_-_138238891
|
0.641
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr13_-_59223074
|
0.641
|
|
|
|
|
chr4_+_114190233
|
0.641
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_+_174084137
|
0.636
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr5_-_138238953
|
0.625
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr17_-_37034288
|
0.624
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr12_-_46405477
|
0.620
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr5_-_32424352
|
0.618
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr6_+_42896771
|
0.610
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr1_-_203657803
|
0.608
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr2_-_160838397
|
0.606
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr17_+_72380323
|
0.600
|
NM_198955
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr7_+_116447527
|
0.600
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr12_-_54392328
|
0.592
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr3_+_191588354
|
0.590
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr14_-_64359565
|
0.589
|
NM_000347
NM_001024858
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr17_-_37022465
|
0.588
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr17_+_36515166
|
0.585
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr8_+_67568044
|
0.584
|
NM_152765
|
C8orf46
|
chromosome 8 open reading frame 46
|
|
chr7_+_30927439
|
0.583
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr18_-_51219880
|
0.582
|
|
TCF4
|
transcription factor 4
|
|
chr11_+_6847499
|
0.582
|
NM_001004460
|
OR10A2
|
olfactory receptor, family 10, subfamily A, member 2
|
|
chr17_+_36493927
|
0.581
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr15_+_87540238
|
0.581
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chrX_-_32340291
|
0.568
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr2_+_157822355
|
0.562
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr21_-_14677310
|
0.561
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr11_-_27700180
|
0.561
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_199519202
|
0.559
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr9_+_35528628
|
0.558
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr13_-_98757634
|
0.557
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr8_+_75899326
|
0.557
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr7_+_116447499
|
0.556
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr2_+_170742900
|
0.554
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr5_+_59819761
|
0.550
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr6_-_56615544
|
0.547
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr3_-_12561962
|
0.546
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr10_-_105228986
|
0.543
|
NM_001129742
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr17_+_36665161
|
0.542
|
NM_030975
|
KRTAP9-9
|
keratin associated protein 9-9
|
|
chr13_-_98428244
|
0.542
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr6_-_56824371
|
0.538
|
|
DST
|
dystonin
|
|
chr15_-_31147376
|
0.534
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr12_-_131725848
|
0.534
|
|
POLE
|
polymerase (DNA directed), epsilon
|
|
chr8_+_70541412
|
0.532
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr12_-_118725672
|
0.532
|
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr1_-_201420927
|
0.531
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr20_-_22978141
|
0.527
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr11_-_7917642
|
0.523
|
NM_001003745
|
OR10A3
|
olfactory receptor, family 10, subfamily A, member 3
|
|
chr9_-_16717887
|
0.522
|
|
BNC2
|
basonuclin 2
|
|
chr11_-_57947358
|
0.522
|
NM_001005566
|
OR5B2
|
olfactory receptor, family 5, subfamily B, member 2
|
|
chr7_+_110518297
|
0.519
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr1_-_152445432
|
0.514
|
NM_001010979
|
C1orf189
|
chromosome 1 open reading frame 189
|
|
chr10_-_90601655
|
0.512
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr1_-_157772420
|
0.511
|
NM_001004469
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5
|
|
chr8_+_12847521
|
0.509
|
NM_020844
|
KIAA1456
|
KIAA1456
|
|
chr12_+_99712504
|
0.509
|
NM_178826
|
ANO4
|
anoctamin 4
|
|
chr11_-_8788765
|
0.507
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr11_+_123625632
|
0.506
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr12_+_7039453
|
0.502
|
|
|
|
|
chr17_+_35484204
|
0.501
|
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr19_+_15699833
|
0.501
|
NM_013939
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2
|
|
chr9_-_35962454
|
0.499
|
|
YBX1
|
Y box binding protein 1
|
|
chr7_+_143263191
|
0.497
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chrX_-_135166306
|
0.495
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr11_-_57173823
|
0.494
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr8_-_37576515
|
0.494
|
|
LOC157860
|
hypothetical protein LOC157860
|
|
chr10_-_23673777
|
0.488
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr2_-_40510923
|
0.485
|
NM_001112800
NM_001112801
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr20_+_2744947
|
0.483
|
NM_001167670
|
LOC100288797
|
hypothetical protein LOC100288797
|
|
chr2_+_105224631
|
0.482
|
NM_007227
|
GPR45
|
G protein-coupled receptor 45
|
|
chr11_-_57171554
|
0.482
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_-_48576204
|
0.482
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr4_-_5881271
|
0.480
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr15_-_81037554
|
0.480
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr1_+_201711505
|
0.478
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr10_+_18280773
|
0.476
|
NM_001145195
NM_152725
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chr7_-_6441109
|
0.474
|
|
DAGLB
|
diacylglycerol lipase, beta
|
|
chr13_+_97403929
|
0.472
|
NM_002271
|
IPO5
|
importin 5
|
|
chr9_-_132759045
|
0.471
|
NM_198180
|
QRFP
|
pyroglutamylated RFamide peptide
|
|
chr14_+_20537253
|
0.469
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr4_+_72423633
|
0.469
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr10_-_92670764
|
0.469
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr1_+_165330710
|
0.468
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr1_+_213245507
|
0.467
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr14_+_104244998
|
0.467
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr16_+_15503623
|
0.465
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr22_-_35026950
|
0.462
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr4_+_86968069
|
0.461
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_-_8636958
|
0.460
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr11_-_123758448
|
0.460
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr3_-_46224791
|
0.459
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr9_+_119506274
|
0.459
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr3_+_25445071
|
0.458
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr5_-_141981083
|
0.453
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_-_107851593
|
0.453
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr12_-_52187638
|
0.452
|
NM_003717
|
NPFF
|
neuropeptide FF-amide peptide precursor
|