|
chr5_+_156628929
|
3.190
|
NM_014376
|
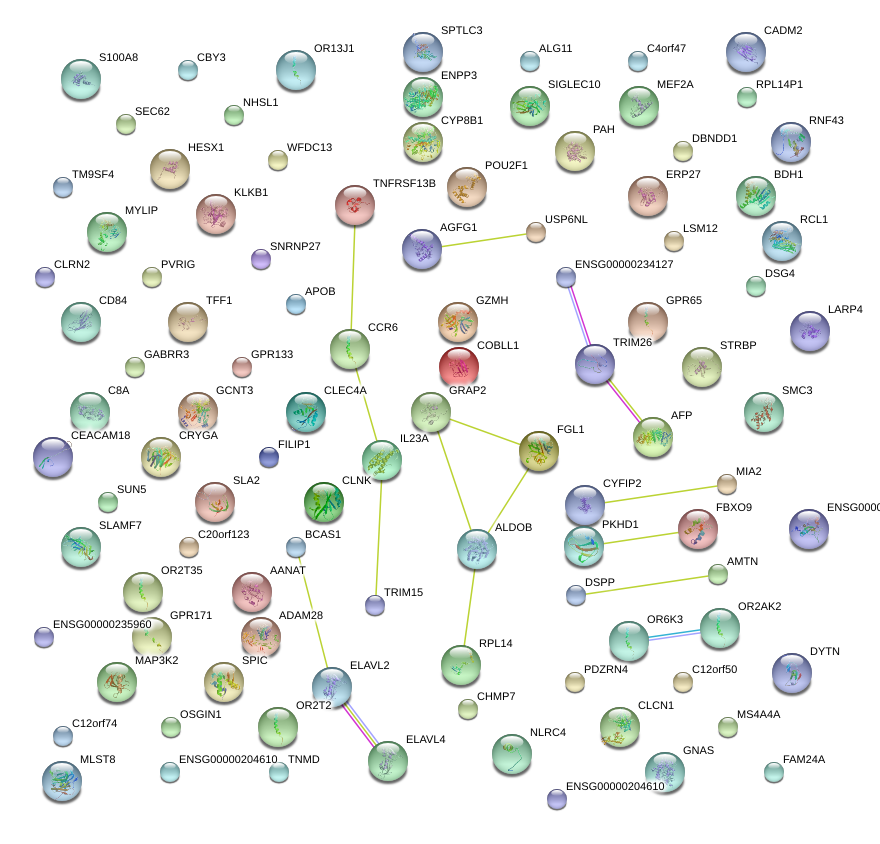
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr20_+_30160952
|
2.385
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr22_+_38652555
|
2.199
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr19_-_56612653
|
2.128
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr14_+_87541220
|
2.060
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr6_+_132000134
|
2.046
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr17_-_53847937
|
1.812
|
|
RNF43
|
ring finger protein 43
|
|
chr17_+_71975224
|
1.637
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr6_-_52060327
|
1.577
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr20_-_52120710
|
1.549
|
NM_003657
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr3_-_42892195
|
1.496
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr1_-_246869181
|
1.414
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr3_-_57209319
|
1.387
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr13_+_51496679
|
1.380
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr9_-_103237851
|
1.349
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr6_+_53043729
|
1.323
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr9_+_4829763
|
1.314
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr6_+_30238961
|
1.303
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr20_+_43764068
|
1.268
|
NM_172005
|
WFDC13
|
WAP four-disulfide core domain 13
|
|
chr2_+_228064531
|
1.236
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr7_+_99654806
|
1.234
|
NM_024070
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing
|
|
chr1_+_246682699
|
1.230
|
NM_001004136
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2
|
|
chr4_+_71418886
|
1.203
|
NM_212557
|
AMTN
|
amelotin
|
|
chr19_+_56671649
|
1.197
|
NM_001080405
|
CEACAM18
|
carcinoembryonic antigen-related cell adhesion molecule 18
|
|
chr2_-_207291364
|
1.196
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr12_+_130004404
|
1.187
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr20_-_34707971
|
1.184
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr20_+_30161173
|
1.183
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr16_+_82540172
|
1.176
|
NM_013370
NM_182980
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr10_-_11614279
|
1.174
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_+_165565006
|
1.058
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr14_-_24148658
|
1.049
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr2_-_32344192
|
1.048
|
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr12_+_91620749
|
1.047
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr3_-_152403636
|
1.036
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr12_+_55018925
|
1.033
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr12_+_8167494
|
1.013
|
NM_016184
NM_194447
NM_194448
NM_194450
|
CLEC4A
|
C-type lectin domain family 4, member A
|
|
chr3_-_198784590
|
1.007
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr18_-_21184824
|
0.990
|
|
|
|
|
chr8_+_23157094
|
0.990
|
NM_152272
|
CHMP7
|
CHMP family, member 7
|
|
chr9_-_124981125
|
0.982
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr2_+_69974581
|
0.977
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr7_+_141457977
|
0.958
|
|
LOC93432
|
maltase-glucoamylase-like pseudogene
|
|
chr1_+_246195157
|
0.942
|
NM_001004491
|
OR2AK2
|
olfactory receptor, family 2, subfamily AK, member 2
|
|
chr2_-_127817143
|
0.931
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr12_+_40117751
|
0.916
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr6_+_167445284
|
0.898
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr12_+_61645361
|
0.884
|
|
RPL14
|
ribosomal protein L14
|
|
chr15_+_97990705
|
0.876
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chrX_+_99726445
|
0.873
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr1_-_151630168
|
0.870
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr3_+_171183203
|
0.866
|
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr21_-_42659671
|
0.858
|
NM_003225
|
TFF1
|
trefoil factor 1
|
|
chr20_+_56919233
|
0.852
|
|
GNAS
|
GNAS complex locus
|
|
chr1_-_156954528
|
0.822
|
NM_001005327
|
OR6K3
|
olfactory receptor, family 6, subfamily K, member 3
|
|
chr1_+_158975662
|
0.817
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chr12_+_100394787
|
0.816
|
NM_152323
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related)
|
|
chr17_+_23818922
|
0.810
|
|
|
|
|
chrX_+_71280758
|
0.786
|
NM_207422
|
FLJ44635
|
TPT1-like protein
|
|
chr9_-_35860316
|
0.763
|
NM_001004487
|
OR13J1
|
olfactory receptor, family 13, subfamily J, member 1
|
|
chr10_+_112334066
|
0.758
|
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr10_+_124660206
|
0.751
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr8_+_24207524
|
0.740
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr4_+_17125885
|
0.739
|
NM_001079827
|
CLRN2
|
clarin 2
|
|
chr2_-_165259334
|
0.735
|
|
COBLL1
|
COBL-like 1
|
|
chr4_-_10295412
|
0.729
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr15_+_57691382
|
0.711
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr12_+_49155798
|
0.698
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr1_-_158815850
|
0.694
|
|
CD84
|
CD84 molecule
|
|
chr20_+_12937616
|
0.693
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr14_+_38772870
|
0.692
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr4_+_187385618
|
0.677
|
NM_000892
|
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1
|
|
chr2_-_21106291
|
0.666
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr4_+_186587537
|
0.661
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr3_+_85858321
|
0.639
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr12_-_101835510
|
0.636
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr5_-_179040580
|
0.634
|
NM_001164444
|
CBY3
|
chibby homolog 3 (Drosophila)
|
|
chr12_-_14982708
|
0.623
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr17_-_16816113
|
0.619
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr20_-_44612619
|
0.613
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr6_-_138862271
|
0.611
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr7_+_142723340
|
0.609
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr18_+_27210737
|
0.609
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr4_+_74520796
|
0.608
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr4_+_88748704
|
0.603
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr1_+_57093030
|
0.603
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr6_-_76259976
|
0.598
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr6_+_16251260
|
0.592
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr3_-_42892636
|
0.586
|
NM_004391
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr17_-_39500512
|
0.585
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr6_-_138862273
|
0.582
|
|
|
|
|
chr16_-_88604029
|
0.577
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr1_+_50348052
|
0.574
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_+_59804634
|
0.573
|
NM_148975
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4
|
|
chr2_-_208736533
|
0.572
|
NM_014617
|
CRYGA
|
crystallin, gamma A
|
|
chr3_-_99236520
|
0.569
|
NM_001105580
|
GABRR3
|
gamma-aminobutyric acid (GABA) receptor, rho 3
|
|
chr8_-_17787342
|
0.567
|
|
FGL1
|
fibrinogen-like 1
|
|
chr12_-_86947276
|
0.567
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr16_+_2195424
|
0.567
|
NM_022372
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae)
|
|
chr20_-_31055892
|
0.557
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr19_-_47638890
|
0.554
|
NM_198477
|
CXCL17
|
chemokine (C-X-C motif) ligand 17
|
|
chr4_+_166350620
|
0.548
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr7_-_36730525
|
0.543
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chrX_-_135783496
|
0.540
|
|
|
|
|
chr1_-_229627412
|
0.539
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr4_+_1872150
|
0.537
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr14_-_74669864
|
0.537
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr6_-_25982418
|
0.527
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr1_+_205329206
|
0.519
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr1_+_559618
|
0.516
|
|
|
|
|
chr1_-_158815895
|
0.511
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr5_-_150663519
|
0.508
|
NM_001145017
NM_181774
|
SLC36A3
|
solute carrier family 36 (proton/amino acid symporter), member 3
|
|
chr1_-_20374273
|
0.501
|
NM_001105572
|
PLA2G2C
|
phospholipase A2, group IIC
|
|
chr20_-_51633042
|
0.501
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr14_+_66901237
|
0.500
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr21_-_30460841
|
0.500
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr12_+_25096472
|
0.499
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr1_-_205012348
|
0.497
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr14_+_38804226
|
0.497
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr4_+_89115862
|
0.491
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr9_+_130056513
|
0.488
|
|
|
|
|
chr6_+_158654464
|
0.486
|
|
TULP4
|
tubby like protein 4
|
|
chr5_+_67622242
|
0.475
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr15_+_57691272
|
0.471
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr8_-_17797127
|
0.462
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr4_-_123761615
|
0.460
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr1_-_20318578
|
0.459
|
NM_012400
|
PLA2G2D
|
phospholipase A2, group IID
|
|
chr8_-_59575249
|
0.455
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr7_+_69869099
|
0.449
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr19_-_3922032
|
0.449
|
|
DAPK3
|
death-associated protein kinase 3
|
|
chr14_+_20493467
|
0.448
|
NM_002934
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin)
|
|
chr11_-_30558495
|
0.448
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr4_+_155703581
|
0.446
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr5_-_160044844
|
0.443
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr17_-_61655915
|
0.442
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr14_+_94148466
|
0.434
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr3_-_120761092
|
0.430
|
NM_005191
|
CD80
|
CD80 molecule
|
|
chr17_-_64752550
|
0.430
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr20_+_43691797
|
0.428
|
NM_080753
|
WFDC10A
|
WAP four-disulfide core domain 10A
|
|
chr6_+_152053323
|
0.428
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr5_+_147672181
|
0.426
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr6_+_64344307
|
0.424
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr6_-_28294642
|
0.422
|
|
LOC222699
|
transducer of ERBB2, 2 pseudogene
|
|
chr11_-_55942283
|
0.418
|
NM_001004744
|
OR5R1
|
olfactory receptor, family 5, subfamily R, member 1
|
|
chr13_+_52500830
|
0.417
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr10_-_69736600
|
0.416
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr6_-_155818728
|
0.416
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr3_+_153074118
|
0.414
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr14_+_88130482
|
0.413
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr4_-_103464491
|
0.406
|
NM_001135148
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8
|
|
chr4_+_89115892
|
0.406
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr17_+_58440629
|
0.406
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr22_-_22426551
|
0.406
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr1_+_66230977
|
0.404
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_-_149681265
|
0.400
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr3_+_187866464
|
0.399
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr2_-_191580567
|
0.397
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr1_-_47179681
|
0.395
|
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11
|
|
chr11_+_58888460
|
0.391
|
NM_001004729
|
OR5AN1
|
olfactory receptor, family 5, subfamily AN, member 1
|
|
chr1_+_114295289
|
0.389
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr5_-_39238818
|
0.389
|
|
FYB
|
FYN binding protein
|
|
chr15_-_74941339
|
0.386
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr3_+_123279439
|
0.386
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr4_-_186969041
|
0.385
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_-_131401319
|
0.384
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr18_-_22696532
|
0.383
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr11_+_67133502
|
0.382
|
|
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa
|
|
chr1_-_157014048
|
0.381
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chr14_-_101617680
|
0.381
|
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1
|
|
chr21_-_26175068
|
0.371
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr19_-_14771947
|
0.369
|
NM_198944
|
OR7C1
|
olfactory receptor, family 7, subfamily C, member 1
|
|
chr20_+_17622319
|
0.369
|
NM_001159495
|
BANF2
|
barrier to autointegration factor 2
|
|
chr1_-_47179729
|
0.368
|
NM_000778
|
CYP4A11
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 11
cytochrome P450, family 4, subfamily A, polypeptide 22
|
|
chr18_-_3209846
|
0.367
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr3_-_156904690
|
0.364
|
NM_014996
|
PLCH1
|
phospholipase C, eta 1
|
|
chr2_-_20375497
|
0.364
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr10_-_98021144
|
0.363
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chrX_+_105823723
|
0.361
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr5_+_180726874
|
0.358
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr12_+_52126131
|
0.358
|
|
PRR13
|
proline rich 13
|
|
chr1_-_180822686
|
0.356
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr9_+_126158935
|
0.355
|
|
|
|
|
chr16_+_21603250
|
0.354
|
NM_001161683
|
OTOA
|
otoancorin
|
|
chr17_-_5078773
|
0.354
|
NM_207103
|
C17orf87
|
chromosome 17 open reading frame 87
|
|
chr6_+_158654890
|
0.352
|
|
|
|
|
chr1_+_151051070
|
0.352
|
NM_178349
|
LCE1B
|
late cornified envelope 1B
|
|
chr12_+_111047676
|
0.351
|
NM_001143906
NM_006700
|
TRAFD1
|
TRAF-type zinc finger domain containing 1
|
|
chrX_+_38096679
|
0.350
|
NM_000531
|
OTC
|
ornithine carbamoyltransferase
|
|
chr12_+_45896511
|
0.349
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr11_+_55335418
|
0.347
|
NM_001004738
|
OR5L1
|
olfactory receptor, family 5, subfamily L, member 1
|
|
chr18_-_22699650
|
0.347
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr2_+_24660849
|
0.346
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr11_+_55362802
|
0.345
|
NM_001005496
|
OR5D16
|
olfactory receptor, family 5, subfamily D, member 16
|
|
chr8_-_124818754
|
0.344
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr2_+_169465995
|
0.342
|
NM_001081686
NM_021176
|
G6PC2
|
glucose-6-phosphatase, catalytic, 2
|
|
chr4_+_75393053
|
0.341
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr14_+_22859236
|
0.341
|
NM_004643
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr14_+_23844269
|
0.341
|
|
|
|
|
chr6_-_137536412
|
0.340
|
NM_052962
NM_181309
NM_181310
|
IL22RA2
|
interleukin 22 receptor, alpha 2
|
|
chr9_-_21218132
|
0.336
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|