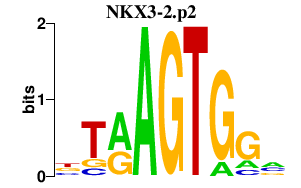
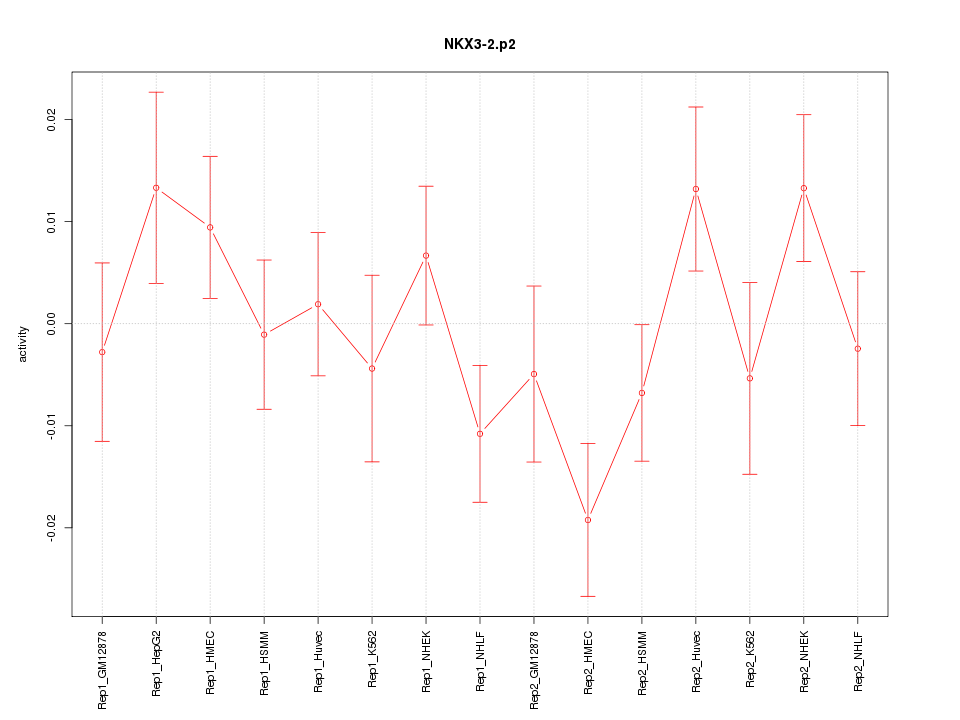
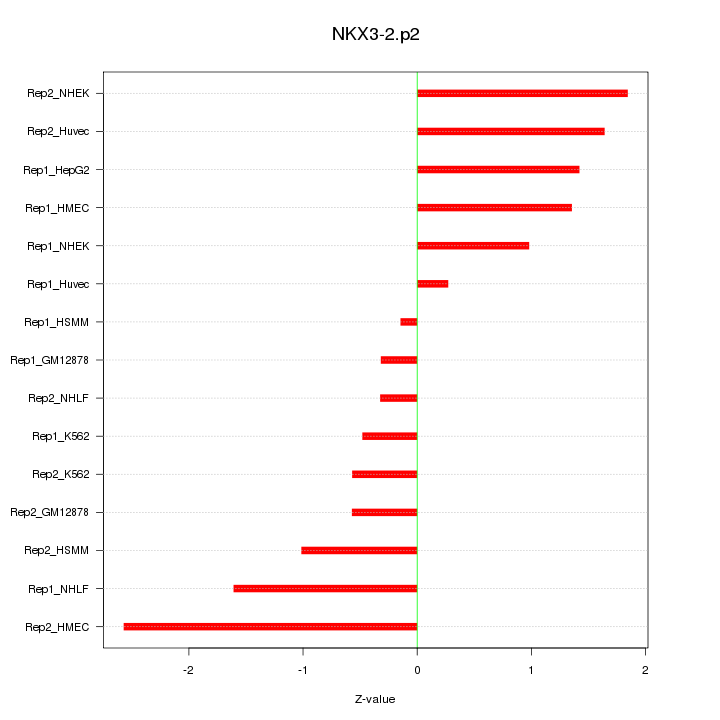
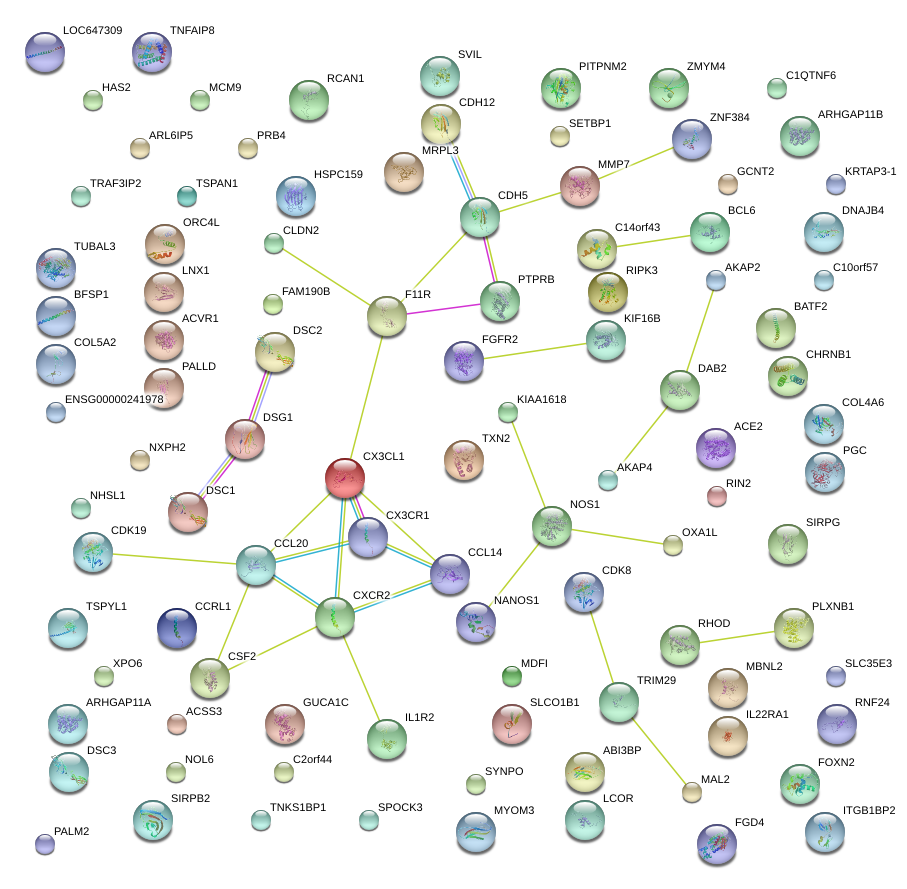
|
chr10_+_82163568
|
1.468
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr2_+_228386800
|
0.918
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr10_-_123343761
|
0.861
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr8_-_122722674
|
0.805
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr6_-_112001590
|
0.751
|
NM_001164282
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr16_-_28130201
|
0.690
|
|
XPO6
|
exportin 6
|
|
chr10_-_5436781
|
0.683
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr2_-_148495642
|
0.627
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr3_+_69216747
|
0.619
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr1_+_46418786
|
0.604
|
|
TSPAN1
|
tetraspanin 1
|
|
chr17_+_71154240
|
0.581
|
NM_001162997
|
LOC100130933
|
hypothetical protein LOC100130933
|
|
chr12_-_122155998
|
0.581
|
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr1_+_35507197
|
0.575
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr2_+_64540843
|
0.574
|
|
HSPC159
|
galectin-related protein
|
|
chr1_-_24311212
|
0.571
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr9_-_33455191
|
0.550
|
|
NOL6
|
nucleolar protein family 6 (RNA-associated)
|
|
chr12_+_32578513
|
0.547
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr20_-_1420004
|
0.506
|
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr14_-_22840249
|
0.503
|
|
|
|
|
chr20_+_19903778
|
0.498
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr5_+_118632403
|
0.497
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chrX_-_107568211
|
0.495
|
NM_033641
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr18_-_26996742
|
0.494
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr5_-_39460787
|
0.487
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr2_-_158439868
|
0.484
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr2_+_101981888
|
0.470
|
NM_173343
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr12_+_32546243
|
0.469
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chrX_-_49852397
|
0.468
|
NM_003886
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr17_-_31337798
|
0.467
|
NM_032962
NM_032963
|
CCL14
|
chemokine (C-C motif) ligand 14
|
|
chr3_-_188936941
|
0.466
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr16_+_64958060
|
0.465
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr2_-_139254273
|
0.442
|
NM_007226
|
NXPH2
|
neurexophilin 2
|
|
chr5_+_118632316
|
0.440
|
NM_001077654
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chr13_+_25726238
|
0.430
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr16_+_64958086
|
0.430
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr3_-_110155237
|
0.422
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr5_-_39460687
|
0.407
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr20_-_1420168
|
0.405
|
NM_001122962
NM_001134836
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr14_-_23878871
|
0.396
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr18_+_40514772
|
0.391
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chrX_+_106050261
|
0.388
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr17_-_36418869
|
0.388
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr3_-_132704339
|
0.384
|
NM_007208
|
MRPL3
|
mitochondrial ribosomal protein L3
|
|
chr15_+_28706170
|
0.378
|
NM_001039841
|
ARHGAP11B
|
Rho GTPase activating protein 11B
|
|
chr6_-_41823056
|
0.377
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr10_+_86174665
|
0.377
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr4_+_169789327
|
0.377
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr20_-_1420126
|
0.373
|
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chrX_+_70438352
|
0.371
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr1_+_35507145
|
0.370
|
NM_005095
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr6_-_138935359
|
0.369
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr1_+_78243181
|
0.366
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chrX_-_114703273
|
0.363
|
|
|
|
|
chr8_+_120289735
|
0.358
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr10_-_29963906
|
0.358
|
NM_021738
|
SVIL
|
supervillin
|
|
chr14_+_22305570
|
0.357
|
NM_005015
|
OXA1L
|
oxidase (cytochrome c) assembly 1-like
|
|
chr11_-_101906640
|
0.357
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chrX_-_15530112
|
0.356
|
NM_021804
|
ACE2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr3_-_102194938
|
0.354
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr10_+_98582648
|
0.354
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr20_-_16501911
|
0.351
|
NM_024704
|
KIF16B
|
kinesin family member 16B
|
|
chr11_-_119514058
|
0.348
|
NM_012101
|
TRIM29
|
tripartite motif containing 29
|
|
chr6_+_10629550
|
0.347
|
NM_145649
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr3_+_133798770
|
0.345
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr5_+_131437383
|
0.344
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr4_-_168392151
|
0.343
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr4_-_168392266
|
0.340
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr6_+_41712907
|
0.339
|
|
MDFI
|
MyoD family inhibitor
|
|
chr13_+_96726197
|
0.338
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr6_-_116706828
|
0.338
|
|
TSPYL1
|
TSPY-like 1
|
|
chr6_-_119297851
|
0.336
|
|
MCM9
|
minichromosome maintenance complex component 9
|
|
chr12_-_116283793
|
0.334
|
NM_000620
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr18_+_27152049
|
0.334
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr22_-_35207582
|
0.332
|
NM_012473
|
TXN2
|
thioredoxin 2
|
|
chr2_+_218698980
|
0.328
|
NM_001557
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr16_+_55963883
|
0.327
|
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_-_39461054
|
0.326
|
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr11_-_56846246
|
0.325
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr2_-_189752575
|
0.324
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_+_67426225
|
0.322
|
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr4_-_54119102
|
0.321
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr20_-_3944035
|
0.319
|
NM_007219
NM_001134337
NM_001134338
|
RNF24
|
ring finger protein 24
|
|
chr16_+_55963899
|
0.319
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr5_-_22248630
|
0.317
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr16_+_64958025
|
0.313
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr11_-_119513948
|
0.313
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr3_-_192063158
|
0.313
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr17_+_7289086
|
0.312
|
NM_000747
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr2_+_48395271
|
0.311
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr12_-_69317441
|
0.311
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr9_+_111582317
|
0.308
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr12_-_6668085
|
0.306
|
NM_001039916
|
ZNF384
|
zinc finger protein 384
|
|
chr14_-_73296387
|
0.302
|
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr20_-_17487480
|
0.302
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr12_+_79995927
|
0.301
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr3_-_48446405
|
0.298
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr5_+_149960820
|
0.296
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr11_+_65023218
|
0.296
|
|
|
|
|
chr22_-_35914200
|
0.295
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr11_+_66580861
|
0.293
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr1_-_24342158
|
0.292
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr11_-_119499188
|
0.292
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr6_-_116529101
|
0.291
|
|
LOC100287673
|
hypothetical LOC100287673
|
|
chr2_-_24123726
|
0.289
|
NM_001142319
NM_025203
|
C2orf44
|
chromosome 2 open reading frame 44
|
|
chr1_-_159257595
|
0.287
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr3_-_123036526
|
0.286
|
|
IQCB1
|
IQ motif containing B1
|
|
chr1_-_23567084
|
0.286
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr6_+_131190237
|
0.286
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr14_-_20636909
|
0.285
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr14_-_49848538
|
0.285
|
NM_024884
|
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase
|
|
chr3_+_113180517
|
0.285
|
NM_018394
|
ABHD10
|
abhydrolase domain containing 10
|
|
chr12_-_121767272
|
0.284
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr14_-_64479198
|
0.282
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr10_+_102113410
|
0.281
|
|
|
|
|
chr4_-_54119123
|
0.280
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr17_+_42023353
|
0.280
|
NM_006178
|
NSF
|
N-ethylmaleimide-sensitive factor
|
|
chr11_-_119514011
|
0.280
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr5_+_66160359
|
0.278
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr14_+_20636935
|
0.278
|
NM_001146683
|
C14orf176
|
chromosome 14 open reading frame 176
|
|
chr4_+_141397889
|
0.274
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr1_-_151630168
|
0.273
|
NM_002964
|
S100A8
|
S100 calcium binding protein A8
|
|
chr6_+_116956868
|
0.271
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr2_-_3360467
|
0.270
|
NM_003310
|
TSSC1
|
tumor suppressing subtransferable candidate 1
|
|
chr7_+_29200636
|
0.270
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr10_-_123343320
|
0.270
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_+_201099093
|
0.269
|
NM_001160033
NM_001160046
NM_152524
|
SGOL2
|
shugoshin-like 2 (S. pombe)
|
|
chr16_+_18903009
|
0.268
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr4_-_168392307
|
0.267
|
NM_001040159
NM_016950
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr9_-_101621964
|
0.267
|
|
|
|
|
chr17_-_6958282
|
0.264
|
NM_080914
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr11_-_64484184
|
0.264
|
NM_001037225
|
C11orf85
|
chromosome 11 open reading frame 85
|
|
chr1_-_183392442
|
0.263
|
NM_030934
|
TRMT1L
|
TRM1 tRNA methyltransferase 1-like
|
|
chr14_-_22610610
|
0.263
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr15_+_62539831
|
0.262
|
|
ZNF609
|
zinc finger protein 609
|
|
chr2_-_68952006
|
0.262
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr1_-_20122638
|
0.261
|
NM_014589
|
PLA2G2E
|
phospholipase A2, group IIE
|
|
chr8_+_32905027
|
0.260
|
|
|
|
|
chrX_-_15198093
|
0.260
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr1_+_15955719
|
0.260
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr18_+_27425727
|
0.258
|
NM_000371
|
TTR
|
transthyretin
|
|
chr12_+_55018925
|
0.257
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr7_+_134114701
|
0.257
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr20_+_35755865
|
0.257
|
|
CTNNBL1
|
catenin, beta like 1
|
|
chr7_-_43875597
|
0.256
|
NM_032014
|
MRPS24
|
mitochondrial ribosomal protein S24
|
|
chr1_+_198263352
|
0.256
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr1_-_11029737
|
0.256
|
NM_006610
NM_139208
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr9_-_27563444
|
0.256
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr5_+_56507029
|
0.256
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr10_+_59942874
|
0.254
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr7_-_50596238
|
0.254
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr4_+_74497998
|
0.253
|
|
ALB
|
albumin
|
|
chr4_+_74498042
|
0.251
|
|
ALB
|
albumin
|
|
chr8_+_22266256
|
0.249
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr16_-_31054563
|
0.249
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr2_-_104834344
|
0.249
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr10_+_96295511
|
0.248
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr5_+_118434966
|
0.247
|
NM_005509
|
DMXL1
|
Dmx-like 1
|
|
chr3_+_62279707
|
0.247
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr1_+_86662356
|
0.247
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr14_+_30564433
|
0.246
|
NM_001128126
NM_007077
|
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit
|
|
chr4_+_166348219
|
0.246
|
NM_007246
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr17_-_40843530
|
0.246
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr16_-_45562840
|
0.245
|
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr14_-_22515725
|
0.244
|
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr6_+_158358067
|
0.244
|
NM_001178088
|
SYNJ2
|
synaptojanin 2
|
|
chrX_+_48545230
|
0.243
|
|
HDAC6
|
histone deacetylase 6
|
|
chr2_-_178645718
|
0.241
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr3_-_79899748
|
0.241
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr3_-_123036597
|
0.240
|
NM_001023570
NM_001023571
|
IQCB1
|
IQ motif containing B1
|
|
chr12_+_49444509
|
0.239
|
|
ATF1
|
activating transcription factor 1
|
|
chr14_-_34414277
|
0.239
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr2_+_27705018
|
0.238
|
NM_001145047
NM_001145048
NM_001145049
|
GPN1
|
GPN-loop GTPase 1
|
|
chr8_-_42515774
|
0.238
|
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr11_-_116199145
|
0.237
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr13_-_95503566
|
0.237
|
NM_020121
|
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2
|
|
chr9_-_112010233
|
0.236
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr1_-_31542178
|
0.236
|
NM_004814
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5)
|
|
chr3_-_58538110
|
0.234
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr22_+_36412277
|
0.233
|
NM_024313
|
NOL12
|
nucleolar protein 12
|
|
chr17_-_57023322
|
0.233
|
NM_199290
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2
|
|
chr4_-_54119159
|
0.232
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr17_-_40348378
|
0.232
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr17_-_39651189
|
0.232
|
NM_014233
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr4_+_35959638
|
0.231
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr6_+_27464461
|
0.230
|
NM_001076781
|
ZNF391
|
zinc finger protein 391
|
|
chr8_+_42515873
|
0.230
|
NM_138436
NM_001135674
NM_001135675
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr14_-_34943574
|
0.229
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr3_-_81893308
|
0.229
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr20_-_49852353
|
0.229
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr14_-_22610572
|
0.229
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr6_+_116528663
|
0.229
|
NM_152729
|
NT5DC1
|
5'-nucleotidase domain containing 1
|
|
chr20_-_581808
|
0.226
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr6_+_139391511
|
0.226
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr14_+_94097513
|
0.225
|
NM_006215
|
SERPINA4
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4
|
|
chr10_+_5126613
|
0.224
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr3_-_191522886
|
0.224
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr3_+_106568243
|
0.224
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr6_+_160068135
|
0.223
|
NM_004906
NM_152858
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr1_-_153209625
|
0.223
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr20_+_51023159
|
0.222
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|