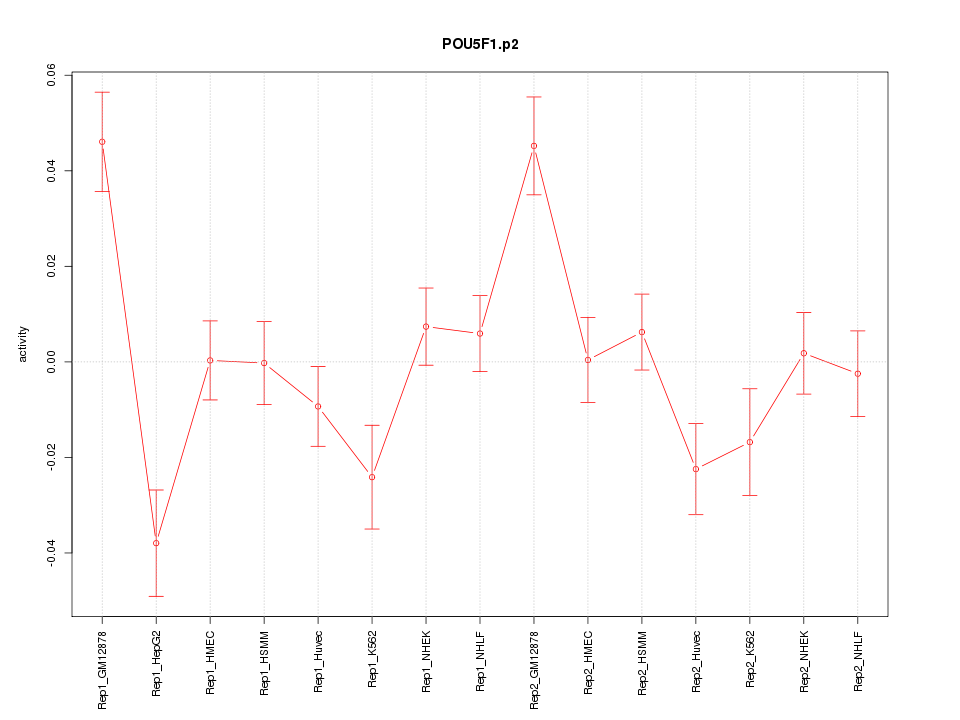
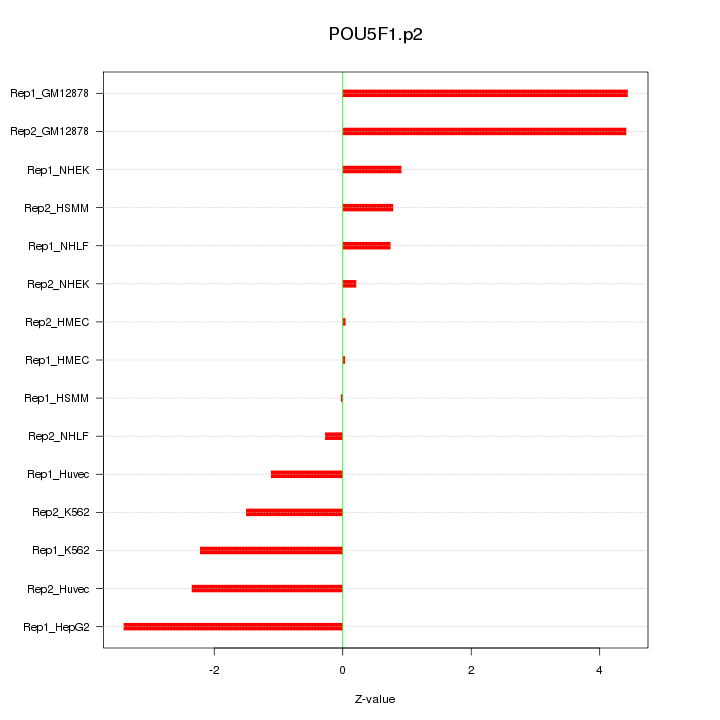
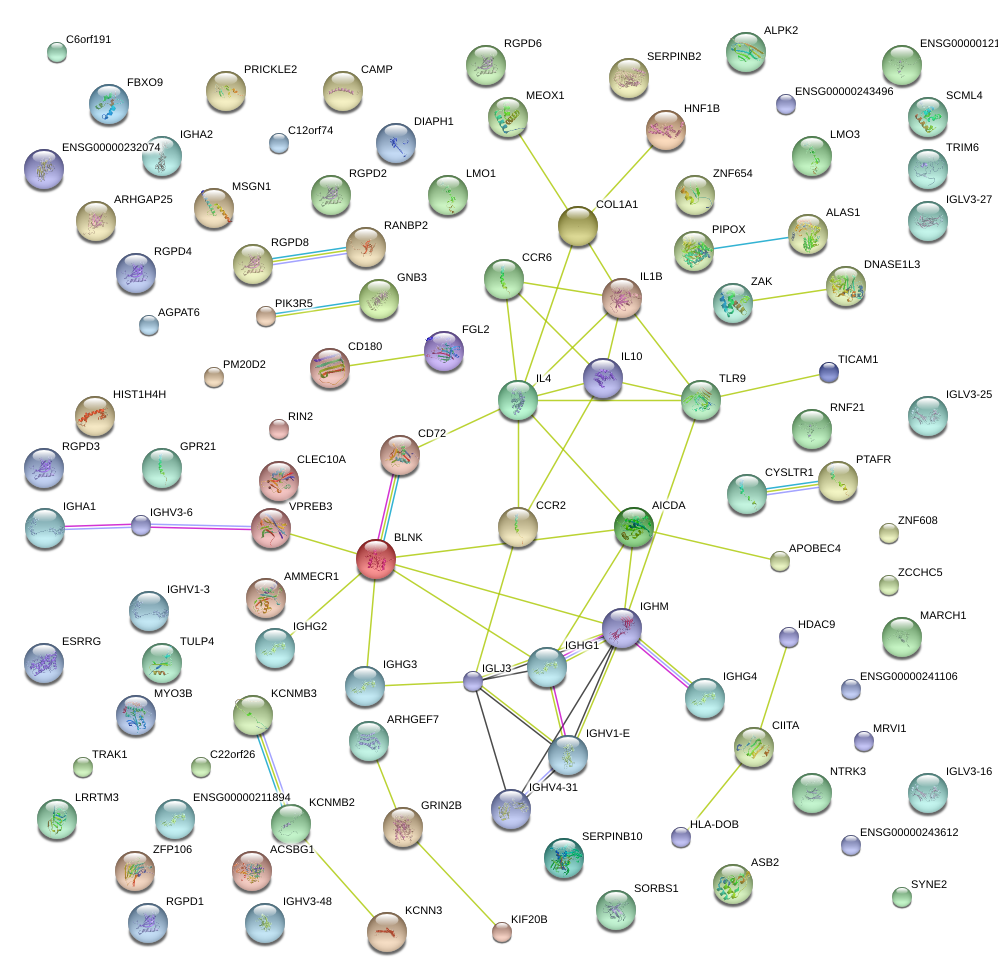
|
chr17_-_8756524
|
5.044
|
NM_014308
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr7_-_76667045
|
4.600
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr14_-_93512818
|
3.949
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr5_+_140871107
|
3.734
|
|
|
|
|
chr2_+_17861266
|
3.732
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr2_+_68815416
|
3.629
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr1_-_181889070
|
3.439
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr17_-_33179118
|
3.299
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_-_153098811
|
3.216
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr14_-_93512647
|
3.182
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr10_-_98021144
|
3.101
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_170742900
|
3.030
|
NM_001083615
NM_001171642
NM_138995
|
MYO3B
|
myosin IIIB
|
|
chr4_-_164754106
|
2.923
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr17_-_33178987
|
2.803
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr3_+_48239865
|
2.663
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr3_+_46370228
|
2.588
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr22_+_21359175
|
2.537
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr15_-_40537002
|
2.499
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr6_+_167456230
|
2.487
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr17_+_24394281
|
2.482
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr3_+_46370578
|
2.384
|
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr14_-_106249999
|
2.318
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr5_-_140875591
|
2.256
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chrX_-_77801480
|
2.215
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr9_+_124836666
|
2.192
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr10_-_97190885
|
2.190
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr9_-_35608300
|
2.179
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr12_+_91620749
|
2.175
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr5_-_66528330
|
2.166
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr14_-_106240998
|
2.148
|
|
IGHV1-69
IGHG1
|
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_-_108252200
|
2.139
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr3_-_52235218
|
2.138
|
NM_017442
|
TLR9
|
toll-like receptor 9
|
|
chr14_-_106241445
|
2.098
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_205012348
|
2.059
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr5_+_140870881
|
2.052
|
|
|
|
|
chr6_+_53043729
|
2.040
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr16_+_10878539
|
1.994
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr7_+_18501876
|
1.934
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr2_+_173794692
|
1.922
|
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr3_-_64186151
|
1.899
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr3_-_58175437
|
1.891
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr14_-_106064887
|
1.833
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr10_+_68355797
|
1.808
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr12_+_6819635
|
1.798
|
NM_002075
|
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3
|
|
chr12_-_16649211
|
1.768
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr22_-_22426551
|
1.755
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr13_+_110637173
|
1.724
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr2_+_108751033
|
1.713
|
|
RANBP2
|
RAN binding protein 2
|
|
chr1_-_215329569
|
1.711
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_+_91488704
|
1.700
|
|
KIF20B
|
kinesin family member 20B
|
|
chr15_-_86600658
|
1.696
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr14_+_63752424
|
1.691
|
NM_182910
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr14_-_106154332
|
1.659
|
|
LOC100126583
IGHG1
|
hypothetical LOC100126583
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_106119742
|
1.656
|
|
IGLJ3
IGHG1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_105804211
|
1.656
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_+_5610060
|
1.644
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr18_+_59726183
|
1.641
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr17_-_39094787
|
1.632
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chrX_-_109570113
|
1.623
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr5_-_124108672
|
1.611
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr18_-_54447168
|
1.611
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr2_+_88966170
|
1.605
|
|
|
|
|
chr3_+_42107711
|
1.596
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chrX_-_77469635
|
1.580
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr1_-_28376041
|
1.577
|
NM_000952
|
PTAFR
|
platelet-activating factor receptor
|
|
chr11_-_10630408
|
1.566
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr14_-_106352471
|
1.537
|
|
|
|
|
chr15_-_76313896
|
1.495
|
NM_015162
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr5_+_132037271
|
1.485
|
NM_000589
NM_172348
|
IL4
|
interleukin 4
|
|
chr17_-_45617899
|
1.461
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr17_+_24394043
|
1.460
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr22_+_20715340
|
1.460
|
|
|
|
|
chr22_+_21495273
|
1.451
|
|
|
|
|
chr6_+_158654464
|
1.441
|
|
TULP4
|
tubby like protein 4
|
|
chr6_-_130224108
|
1.433
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr17_+_45617676
|
1.428
|
|
|
|
|
chr2_-_113310796
|
1.419
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr20_+_19863716
|
1.415
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr22_+_21340757
|
1.414
|
|
|
|
|
chr3_+_88270951
|
1.410
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr3_+_52212969
|
1.408
|
|
ALAS1
|
aminolevulinate, delta-, synthase 1
|
|
chr12_-_14024187
|
1.401
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr12_-_8656705
|
1.391
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr22_-_44828636
|
1.387
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr3_+_179736917
|
1.368
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr6_-_32892705
|
1.347
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr1_+_190552744
|
1.334
|
NM_001039152
|
RGS21
|
regulator of G-protein signaling 21
|
|
chr6_+_26291936
|
1.333
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr19_+_14554895
|
1.331
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr1_-_42156957
|
1.330
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr6_+_158654890
|
1.325
|
|
|
|
|
chr3_-_152478843
|
1.319
|
NM_014879
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr6_+_52159143
|
1.304
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chrX_+_16051344
|
1.288
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr2_+_65517316
|
1.287
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr1_+_208008517
|
1.284
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr6_-_133120587
|
1.280
|
NM_004665
|
VNN2
|
vanin 2
|
|
chr1_+_156526182
|
1.276
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr12_-_121767272
|
1.275
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr10_-_116276651
|
1.264
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_27541049
|
1.256
|
NM_001193308
NM_032872
|
SYTL1
|
synaptotagmin-like 1
|
|
chr1_-_156013487
|
1.243
|
NM_030764
|
FCRL2
|
Fc receptor-like 2
|
|
chr5_-_176869943
|
1.237
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr22_+_21094094
|
1.228
|
|
|
|
|
chr16_+_31274020
|
1.226
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr12_-_103846186
|
1.223
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr1_+_27541086
|
1.215
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr12_-_121753795
|
1.214
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr2_-_191580567
|
1.210
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr15_+_62578671
|
1.204
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chr4_+_76700281
|
1.197
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr11_-_114880275
|
1.196
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr21_+_38566298
|
1.192
|
NM_170737
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr17_-_16816113
|
1.188
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr1_-_197173159
|
1.180
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr14_-_106202262
|
1.180
|
|
IGLJ3
IGHA1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
|
|
chr14_-_105524207
|
1.175
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_-_186970366
|
1.175
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_36337756
|
1.175
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chrX_-_106846924
|
1.174
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_116455898
|
1.173
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr9_-_34681135
|
1.161
|
NM_006274
|
CCL19
|
chemokine (C-C motif) ligand 19
|
|
chr1_-_180908639
|
1.160
|
NM_001102450
NM_033345
|
RGS8
|
regulator of G-protein signaling 8
|
|
chr16_+_31274030
|
1.157
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr3_-_113334657
|
1.149
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr4_+_154397975
|
1.145
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr4_-_110443247
|
1.142
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr11_-_26700121
|
1.141
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr3_-_126325132
|
1.131
|
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr12_+_52713098
|
1.128
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr19_+_40511909
|
1.128
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chrX_+_69591639
|
1.125
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr8_+_11388918
|
1.124
|
NM_001715
|
BLK
|
B lymphoid tyrosine kinase
|
|
chr18_-_21184824
|
1.123
|
|
|
|
|
chr11_+_59920062
|
1.121
|
NM_001079692
NM_032597
|
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14
|
|
chr8_-_102872372
|
1.115
|
|
NCALD
|
neurocalcin delta
|
|
chr1_+_154386358
|
1.109
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr13_-_98708549
|
1.109
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr4_+_81325447
|
1.102
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr5_+_49997489
|
1.101
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr1_-_42157077
|
1.084
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr4_-_47534815
|
1.082
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr6_+_37245860
|
1.082
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr22_+_28806452
|
1.076
|
NM_152510
|
HORMAD2
|
HORMA domain containing 2
|
|
chr8_-_95289945
|
1.069
|
NM_004063
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr6_-_32665409
|
1.062
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr16_+_31273969
|
1.056
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr10_+_95507555
|
1.044
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr12_-_13139848
|
1.038
|
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr12_+_14409875
|
1.034
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr1_+_61315378
|
1.033
|
|
|
|
|
chr7_+_28418668
|
1.028
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr22_+_21053972
|
1.024
|
|
|
|
|
chr5_-_156468614
|
1.022
|
NM_032782
|
HAVCR2
|
hepatitis A virus cellular receptor 2
|
|
chrX_+_21960841
|
1.020
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr6_-_31668476
|
1.018
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr9_+_131123115
|
1.018
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr14_-_105589467
|
1.013
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr6_-_41276901
|
1.007
|
NM_024807
|
TREML2
|
triggering receptor expressed on myeloid cells-like 2
|
|
chrX_-_76700873
|
1.006
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr5_-_160211623
|
0.999
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr1_+_190811479
|
0.995
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr7_+_50318801
|
0.986
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_+_117345894
|
0.980
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr17_-_74231240
|
0.971
|
|
CYTH1
|
cytohesin 1
|
|
chr10_-_103593529
|
0.962
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr10_+_95838801
|
0.961
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr14_-_106184988
|
0.958
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_-_36991189
|
0.958
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr7_+_128795178
|
0.958
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr2_-_215605054
|
0.958
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr2_-_191723951
|
0.956
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr12_-_16652202
|
0.952
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_-_123130435
|
0.951
|
NM_001005188
|
OR6X1
|
olfactory receptor, family 6, subfamily X, member 1
|
|
chr6_+_131613196
|
0.950
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr1_+_200246370
|
0.944
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr7_+_129057152
|
0.939
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr7_+_128795280
|
0.935
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr4_+_124540114
|
0.930
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr22_+_21370394
|
0.926
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr11_+_119616112
|
0.923
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr20_+_34937928
|
0.923
|
NM_080628
|
C20orf118
|
chromosome 20 open reading frame 118
|
|
chr4_-_153523113
|
0.920
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_101398451
|
0.917
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_+_61315516
|
0.912
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_17599438
|
0.910
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr12_-_10498481
|
0.905
|
NM_213657
NM_213658
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr22_-_34886727
|
0.905
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr2_-_191723978
|
0.898
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr10_+_86121526
|
0.891
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr11_-_114593838
|
0.889
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr16_+_68284252
|
0.887
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr14_-_106038122
|
0.887
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr15_-_41879426
|
0.887
|
NM_001033517
|
SERINC4
|
serine incorporator 4
|
|
chrX_+_65152004
|
0.884
|
|
|
|
|
chr1_-_239865724
|
0.880
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr17_+_69781980
|
0.876
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr9_+_115382990
|
0.876
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr8_+_23157094
|
0.875
|
NM_152272
|
CHMP7
|
CHMP family, member 7
|
|
chr19_+_12995782
|
0.875
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|