|
chr1_+_166516901
|
4.798
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr22_+_38652555
|
4.512
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr6_+_7524691
|
4.168
|
|
DSP
|
desmoplakin
|
|
chr4_-_145281237
|
3.892
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr15_-_100280784
|
3.430
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr1_+_158603480
|
3.390
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr4_-_145159889
|
3.296
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr1_+_165565006
|
3.291
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_165564904
|
3.288
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr8_-_13018123
|
3.273
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr11_+_5466490
|
3.061
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr1_+_58899
|
3.016
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr19_-_54350457
|
2.997
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr3_+_129682434
|
2.806
|
|
|
|
|
chr10_+_94584449
|
2.799
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr17_-_36232366
|
2.776
|
NM_000421
|
KRT10
|
keratin 10
|
|
chrX_-_55074125
|
2.640
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr19_+_61624
|
2.621
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr11_+_19328846
|
2.552
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr7_-_92693716
|
2.448
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chrX_+_65299157
|
2.389
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr4_+_71806538
|
2.371
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr6_-_49942147
|
2.303
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr3_-_13032130
|
2.275
|
|
|
|
|
chr11_-_123758448
|
2.187
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr3_+_46894239
|
2.051
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr16_+_33112480
|
1.929
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr1_-_229071957
|
1.920
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr22_+_21116283
|
1.813
|
|
|
|
|
chr9_-_94206648
|
1.806
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr7_-_107667728
|
1.770
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_+_153074118
|
1.721
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr16_+_33169015
|
1.710
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr16_-_32595553
|
1.616
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr9_+_19280748
|
1.607
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr6_+_105511615
|
1.600
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr1_-_158815895
|
1.581
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr6_-_123999640
|
1.570
|
NM_006073
|
TRDN
|
triadin
|
|
chr20_+_56919233
|
1.564
|
|
GNAS
|
GNAS complex locus
|
|
chr7_+_91762227
|
1.547
|
|
|
|
|
chr5_-_75954807
|
1.469
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr6_-_138231006
|
1.465
|
|
|
|
|
chr17_-_10362583
|
1.438
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr19_-_38052481
|
1.407
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr1_+_109057078
|
1.393
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr13_+_108079570
|
1.379
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chr22_+_21065134
|
1.370
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr4_-_44348414
|
1.368
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr2_+_128009782
|
1.364
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr2_-_169596078
|
1.355
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr1_+_46151845
|
1.306
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr11_-_57173823
|
1.286
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr22_+_21065202
|
1.247
|
|
|
|
|
chr11_+_69176391
|
1.238
|
|
CCND1
|
cyclin D1
|
|
chr4_-_145046109
|
1.230
|
NM_002102
NM_198682
|
GYPE
|
glycophorin E (MNS blood group)
|
|
chr1_+_61642287
|
1.218
|
|
NFIA
|
nuclear factor I/A
|
|
chr2_-_180135559
|
1.217
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr2_-_157890400
|
1.215
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr2_-_225073357
|
1.209
|
|
CUL3
|
cullin 3
|
|
chr4_+_39874921
|
1.200
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr19_+_44591589
|
1.188
|
|
|
|
|
chr2_+_233099113
|
1.183
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr13_+_112746908
|
1.178
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr12_-_88573949
|
1.150
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr1_+_559618
|
1.142
|
|
|
|
|
chr13_-_43908937
|
1.133
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr1_-_56750235
|
1.044
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_+_163904564
|
1.023
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr3_-_57209319
|
1.022
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr7_-_135263737
|
1.013
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr20_+_62266217
|
1.003
|
|
MYT1
|
myelin transcription factor 1
|
|
chr7_+_138469029
|
0.984
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr18_+_45342398
|
0.968
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr5_-_39238818
|
0.966
|
|
FYB
|
FYN binding protein
|
|
chr1_-_41399632
|
0.952
|
NM_001031694
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr5_+_140583098
|
0.947
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr5_-_145232723
|
0.936
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr6_-_2696152
|
0.893
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chrX_+_65299352
|
0.888
|
|
HEPH
|
hephaestin
|
|
chr10_+_63331401
|
0.885
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr12_+_108310906
|
0.879
|
NM_001101421
|
MYO1H
|
myosin IH
|
|
chr3_-_153470022
|
0.877
|
|
LOC401093
|
hypothetical LOC401093
|
|
chr11_-_123772456
|
0.872
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr6_-_127822227
|
0.870
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr12_-_16652202
|
0.867
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_+_120490054
|
0.861
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr5_-_86739683
|
0.850
|
|
CCNH
|
cyclin H
|
|
chr18_+_606671
|
0.839
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr22_-_21193504
|
0.829
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr5_+_176449156
|
0.824
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr1_-_161380562
|
0.821
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr20_+_62266270
|
0.818
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr22_+_21065207
|
0.816
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr17_-_36276987
|
0.807
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr1_-_93030538
|
0.802
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr6_+_53991672
|
0.796
|
NM_138569
|
C6orf142
|
chromosome 6 open reading frame 142
|
|
chr2_+_178893216
|
0.794
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr13_-_43908991
|
0.787
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr7_+_72034735
|
0.783
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr13_-_43908985
|
0.775
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_43908995
|
0.772
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr13_-_43908994
|
0.771
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr5_+_139869819
|
0.767
|
|
|
|
|
chr1_+_224079597
|
0.755
|
NM_000120
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr6_+_31604717
|
0.752
|
NM_001011700
|
MCCD1
|
mitochondrial coiled-coil domain 1
|
|
chr16_+_2672527
|
0.747
|
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr15_-_48626193
|
0.741
|
NM_203494
|
USP50
|
ubiquitin specific peptidase 50
|
|
chr7_-_50596238
|
0.726
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr1_+_556968
|
0.723
|
|
|
|
|
chr11_-_16454493
|
0.719
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_-_139960874
|
0.715
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr13_-_43908972
|
0.709
|
NM_006022
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr3_-_197726587
|
0.695
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr7_+_143043871
|
0.684
|
NM_001130026
|
FAM115C
|
family with sequence similarity 115, member C
|
|
chr1_-_158815850
|
0.683
|
|
CD84
|
CD84 molecule
|
|
chr11_+_4965996
|
0.677
|
NM_021801
|
MMP26
|
matrix metallopeptidase 26
|
|
chr3_+_142175329
|
0.674
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr8_-_101384633
|
0.672
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr15_+_47502666
|
0.670
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr7_+_129771865
|
0.670
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr21_+_37714470
|
0.661
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr17_+_46176105
|
0.659
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr3_+_162697289
|
0.636
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr17_-_10265991
|
0.629
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr6_-_27988152
|
0.619
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr10_+_70172257
|
0.618
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr4_-_164614335
|
0.617
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr1_-_120189301
|
0.608
|
NM_001047980
|
NBPF7
|
neuroblastoma breakpoint family, member 7
|
|
chr2_-_105421661
|
0.587
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr17_-_38087366
|
0.586
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr7_+_142539291
|
0.585
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr17_-_64462976
|
0.585
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr1_-_201321711
|
0.585
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr18_-_3209846
|
0.583
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr7_+_128136347
|
0.582
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr14_-_60022487
|
0.581
|
NM_174978
|
C14orf39
|
chromosome 14 open reading frame 39
|
|
chr1_-_178100204
|
0.572
|
|
|
|
|
chr5_+_140509803
|
0.568
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr17_-_53847937
|
0.566
|
|
RNF43
|
ring finger protein 43
|
|
chr11_+_61943082
|
0.557
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr2_+_169367345
|
0.552
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chrX_-_153698930
|
0.552
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr11_+_71605466
|
0.551
|
NM_000803
NM_001113534
NM_001113535
NM_001113536
|
FOLR2
|
folate receptor 2 (fetal)
|
|
chr4_+_42590038
|
0.551
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr3_-_198784590
|
0.544
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr6_-_29163027
|
0.543
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr12_-_69434330
|
0.536
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr17_+_55269974
|
0.532
|
|
|
|
|
chr7_+_106897737
|
0.531
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_-_159543847
|
0.529
|
|
MPZ
|
myelin protein zero
|
|
chr14_-_20128721
|
0.518
|
NM_001024822
|
RNASE12
|
ribonuclease, RNase A family, 12 (non-active)
|
|
chr7_+_18296569
|
0.518
|
|
HDAC9
|
histone deacetylase 9
|
|
chr5_+_86712376
|
0.510
|
|
|
|
|
chr12_-_7739591
|
0.503
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr11_+_86710076
|
0.500
|
|
TMEM135
|
transmembrane protein 135
|
|
chr7_+_38984133
|
0.499
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr13_+_108046500
|
0.493
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr12_-_110410730
|
0.485
|
|
ATXN2
|
ataxin 2
|
|
chr11_+_59564323
|
0.482
|
NM_173801
|
PLAC1L
|
placenta-specific 1-like
|
|
chr16_-_32596378
|
0.478
|
NM_001099687
|
TP53TG3B
|
TP53 target 3B
|
|
chrX_+_123062143
|
0.471
|
|
STAG2
|
stromal antigen 2
|
|
chr8_+_17440664
|
0.470
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr17_+_20712337
|
0.469
|
|
|
|
|
chr22_+_21370394
|
0.468
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr11_+_5329313
|
0.467
|
NM_001004750
|
OR51B6
|
olfactory receptor, family 51, subfamily B, member 6
|
|
chr17_-_57023322
|
0.467
|
NM_199290
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2
|
|
chr6_+_45404031
|
0.467
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr12_-_49897743
|
0.464
|
|
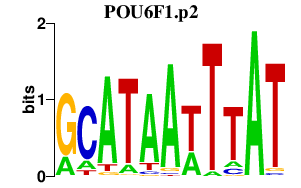
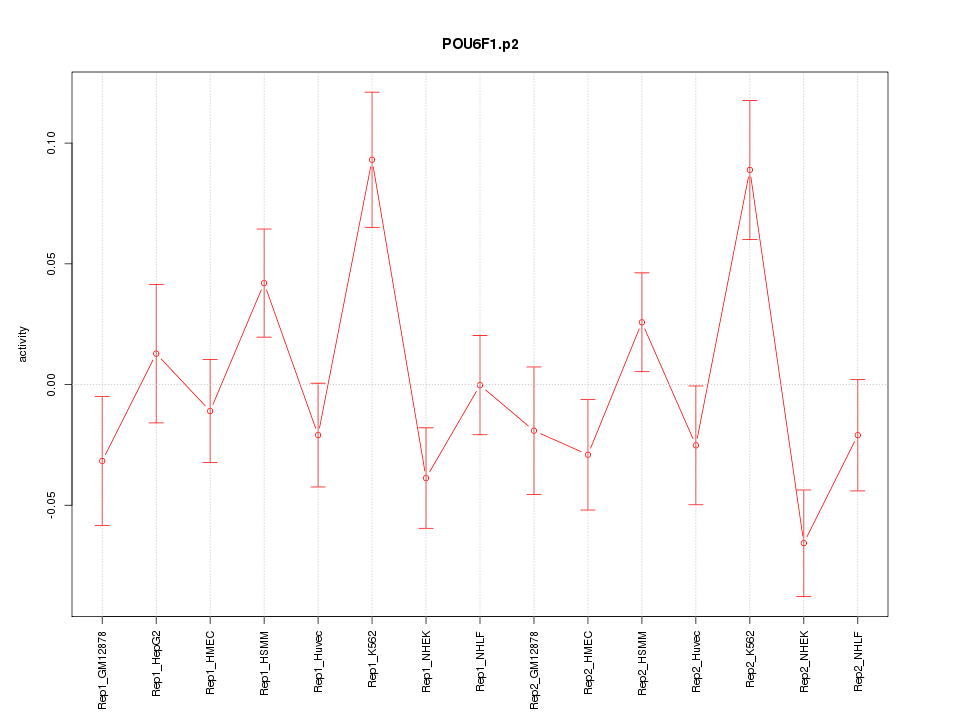
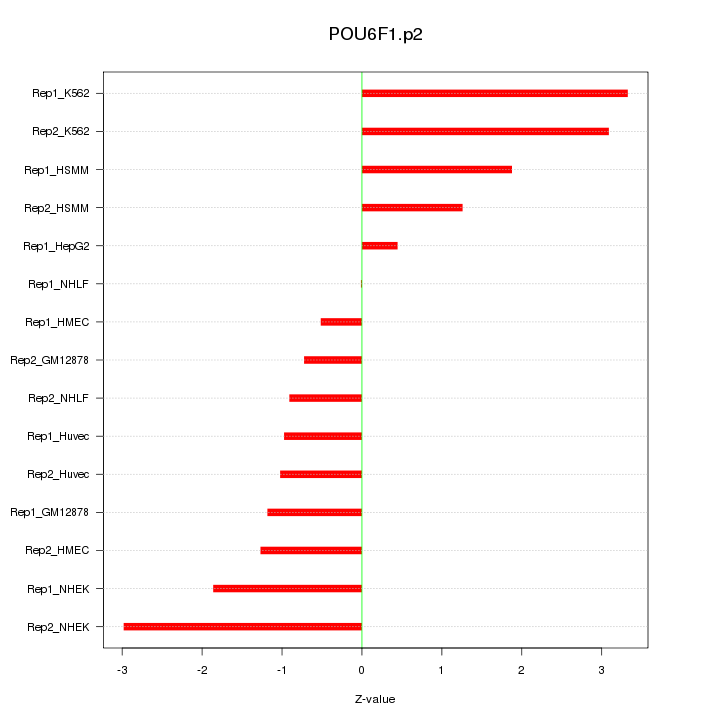
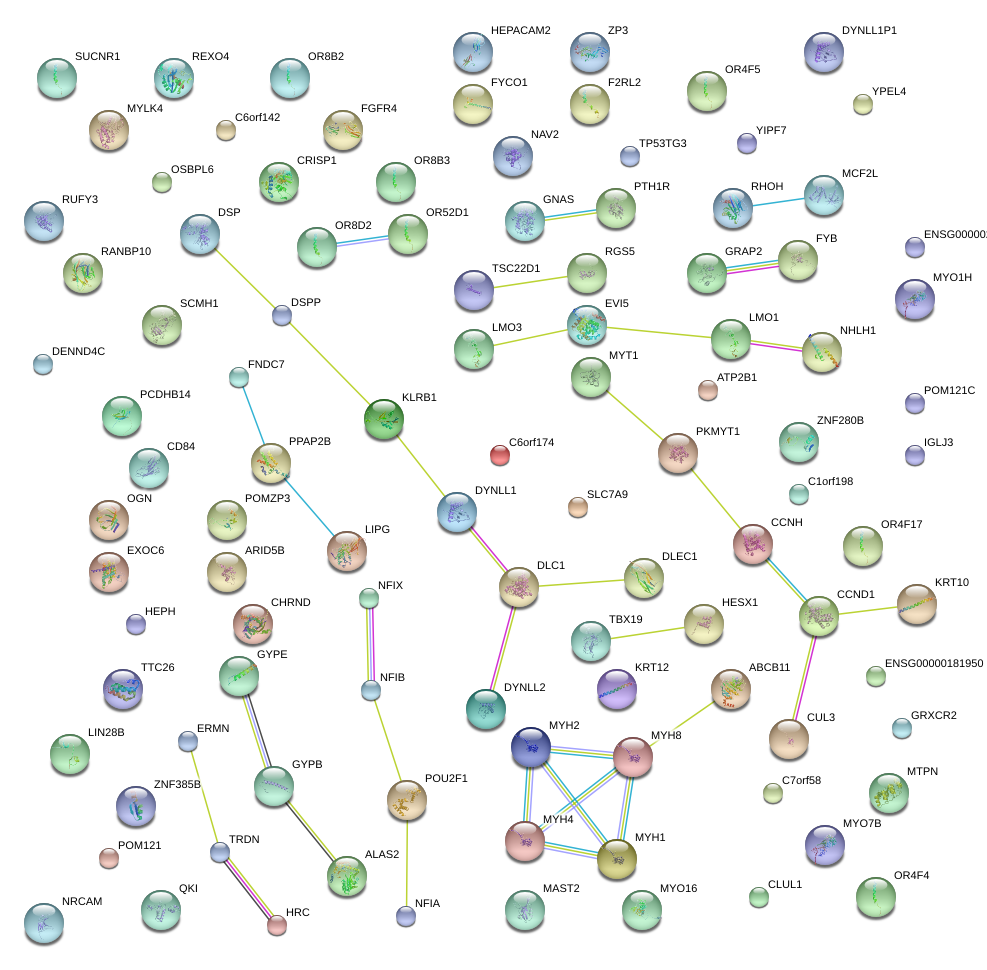
POU6F1
|
POU class 6 homeobox 1
|
|
chr22_+_21042091
|
0.461
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr2_-_215953818
|
0.457
|
|
FN1
|
fibronectin 1
|
|
chr14_+_19473606
|
0.456
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr7_-_25234582
|
0.448
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr4_-_153493133
|
0.445
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr7_+_94130673
|
0.445
|
|
PEG10
|
paternally expressed 10
|
|
chr10_-_13584950
|
0.442
|
NM_001100912
NM_152751
|
BEND7
|
BEN domain containing 7
|
|
chr3_-_114047479
|
0.442
|
NM_001008784
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr16_+_2593351
|
0.442
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr9_+_80134302
|
0.441
|
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr2_-_158439868
|
0.439
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr11_+_77452550
|
0.437
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr12_+_54611212
|
0.435
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr1_+_74473672
|
0.432
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr7_+_104468614
|
0.425
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr3_-_121117647
|
0.424
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr17_-_37794017
|
0.422
|
NM_003150
NM_139276
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr17_-_52248248
|
0.420
|
NM_001085430
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr7_-_112514990
|
0.419
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr6_+_42896771
|
0.417
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr13_-_45577144
|
0.416
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr19_-_59295882
|
0.414
|
NM_130771
NM_133168
NM_133169
NM_206818
|
OSCAR
|
osteoclast associated, immunoglobulin-like receptor
|
|
chr6_-_49862858
|
0.410
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr1_+_196874839
|
0.408
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr6_+_144654565
|
0.408
|
NM_007124
|
UTRN
|
utrophin
|
|
chr1_-_149611768
|
0.406
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr7_-_5533621
|
0.405
|
|
ACTB
|
actin, beta
|
|
chr17_-_10313600
|
0.404
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr22_+_19458378
|
0.401
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr5_+_150020595
|
0.401
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr6_+_29187565
|
0.401
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr7_-_99864220
|
0.396
|
NM_017984
|
ZCWPW1
|
zinc finger, CW type with PWWP domain 1
|