|
chr22_+_35586950
|
5.684
|
NM_000631
NM_013416
|
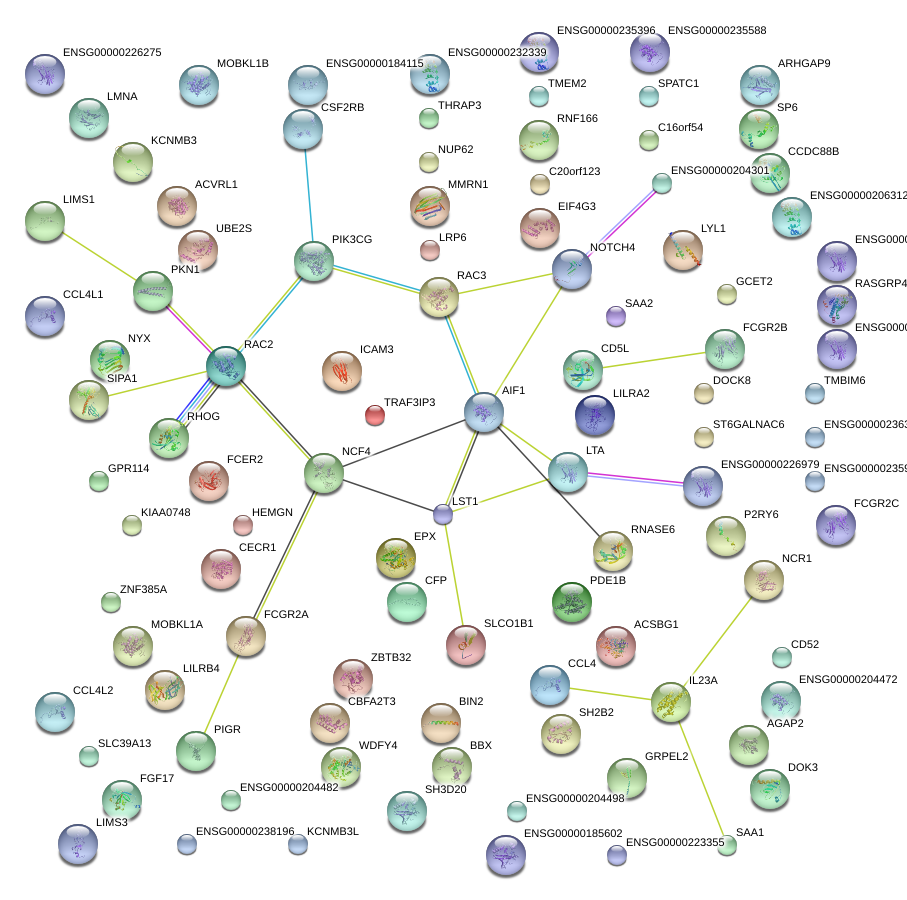
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chrX_-_47374187
|
4.842
|
NM_001145252
|
CFP
|
complement factor properdin
|
|
chr1_+_159899528
|
4.539
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr12_+_48443232
|
4.203
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr16_-_87570716
|
3.690
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chrX_-_47374647
|
3.319
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr1_+_159817752
|
3.119
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr10_+_49562866
|
3.005
|
|
WDFY4
|
WDFY family member 4
|
|
chr6_+_31690957
|
2.999
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr11_+_65164767
|
2.956
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr6_+_31661934
|
2.927
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr6_+_31662454
|
2.809
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_+_108604111
|
2.778
|
NM_001193484
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr19_-_10311262
|
2.636
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr17_+_53625084
|
2.591
|
NM_000502
|
EPX
|
eosinophil peroxidase
|
|
chr11_+_63864263
|
2.547
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr14_+_20319049
|
2.536
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr19_-_46504831
|
2.520
|
|
|
|
|
chr7_+_106292932
|
2.495
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr16_+_28413421
|
2.424
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr16_-_29664765
|
2.363
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr12_+_50592379
|
2.337
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr7_+_106292975
|
2.316
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr9_-_129707392
|
2.258
|
|
LOC100131355
ST6GALNAC6
|
hypothetical protein LOC100131355
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr16_+_56134077
|
2.241
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr17_-_40843530
|
2.197
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr16_+_56134037
|
2.180
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr19_-_55104875
|
2.147
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr19_-_7672978
|
2.138
|
NM_002002
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr17_+_31455332
|
2.092
|
NM_002984
|
CCL4
|
chemokine (C-C motif) ligand 4
|
|
chr3_-_180467483
|
1.987
|
NM_001163677
NM_171828
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr6_+_31648071
|
1.919
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr12_+_55018925
|
1.891
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr19_-_43608641
|
1.885
|
NM_001146202
NM_001146203
NM_001146204
NM_001146205
NM_001146206
NM_001146207
NM_170604
|
RASGRP4
|
RAS guanyl releasing protein 4
|
|
chr12_+_53241365
|
1.834
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr9_+_263025
|
1.828
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr19_-_13074973
|
1.788
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr19_+_59866217
|
1.773
|
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr22_-_16080265
|
1.747
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr17_-_43288137
|
1.742
|
NM_199262
|
SP6
|
Sp6 transcription factor
|
|
chr1_-_21375939
|
1.712
|
NM_001198801
NM_001198802
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr1_-_156078115
|
1.660
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr6_+_31647854
|
1.643
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr7_+_101715110
|
1.643
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr19_+_14412080
|
1.641
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr19_+_14412052
|
1.638
|
|
PKN1
|
protein kinase N1
|
|
chr8_+_145158432
|
1.635
|
NM_001134374
NM_198572
|
SPATC1
|
spermatogenesis and centriole associated 1
|
|
chr22_+_35639564
|
1.625
|
NM_000395
|
CSF2RB
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage)
|
|
chr12_-_12310969
|
1.604
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr19_+_40895669
|
1.600
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr1_+_26517032
|
1.568
|
|
CD52
|
CD52 molecule
|
|
chr1_+_154351200
|
1.549
|
|
LMNA
|
lamin A/C
|
|
chr12_-_56422120
|
1.505
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr5_+_148711419
|
1.478
|
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr22_-_35970230
|
1.475
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr8_+_21956335
|
1.463
|
NM_003867
|
FGF17
|
fibroblast growth factor 17
|
|
chr17_+_31562577
|
1.459
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr3_+_108724472
|
1.454
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr19_+_14412087
|
1.437
|
|
PKN1
|
protein kinase N1
|
|
chr12_-_50004102
|
1.432
|
|
BIN2
|
bridging integrator 2
|
|
chr7_+_101715137
|
1.432
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr12_+_48443588
|
1.425
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr15_-_76313896
|
1.419
|
NM_015162
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1
|
|
chr11_-_18226743
|
1.398
|
NM_001127380
NM_030754
|
SAA2
|
serum amyloid A2
|
|
chr1_+_36462625
|
1.397
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr11_+_47386707
|
1.395
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr20_-_44612619
|
1.394
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chrX_+_41191630
|
1.367
|
NM_022567
|
NYX
|
nyctalopin
|
|
chr11_+_47386621
|
1.354
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr1_+_26517045
|
1.353
|
|
CD52
|
CD52 molecule
|
|
chr19_+_60109319
|
1.352
|
NM_001145457
NM_001145458
NM_004829
|
NCR1
|
natural cytotoxicity triggering receptor 1
|
|
chr12_-_50004208
|
1.352
|
|
BIN2
|
bridging integrator 2
|
|
chr16_-_87297525
|
1.349
|
NM_001171816
|
RNF166
|
ring finger protein 166
|
|
chr1_-_205186330
|
1.334
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr3_-_113334657
|
1.332
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr4_+_71986927
|
1.330
|
NM_173468
|
MOBKL1A
|
MOB1, Mps One Binder kinase activator-like 1A (yeast)
|
|
chr12_-_50004172
|
1.322
|
NM_016293
|
BIN2
|
bridging integrator 2
|
|
chr12_-_56159631
|
1.319
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr12_-_53071285
|
1.319
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr6_-_32299757
|
1.313
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr9_-_99740243
|
1.306
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr19_-_60611136
|
1.289
|
NM_014501
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr22_-_35970133
|
1.284
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr9_-_73573121
|
1.275
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr19_+_59776293
|
1.267
|
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2
|
|
chr19_-_13074669
|
1.266
|
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr12_-_53664722
|
1.258
|
NM_001136030
|
KIAA0748
|
KIAA0748
|
|
chr1_+_207995999
|
1.251
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr6_+_31691745
|
1.236
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr17_+_31664143
|
1.235
|
NM_207007
NM_001001435
|
CCL4L2
CCL4L1
|
chemokine (C-C motif) ligand 4-like 2
chemokine (C-C motif) ligand 4-like 1
|
|
chr11_+_72658586
|
1.230
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr4_+_91035025
|
1.211
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr5_-_176869943
|
1.207
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr5_-_176258920
|
1.207
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr12_-_56159895
|
1.200
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr1_-_145712107
|
1.197
|
NM_005266
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr7_+_149895390
|
1.195
|
NM_018326
|
GIMAP4
|
GTPase, IMAP family member 4
|
|
chr22_+_42900168
|
1.194
|
NM_001137605
|
PARVG
|
parvin, gamma
|
|
chrX_-_44945025
|
1.193
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr2_+_66519949
|
1.188
|
|
MEIS1
|
Meis homeobox 1
|
|
chr17_-_31441390
|
1.183
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr12_-_53065758
|
1.182
|
|
ZNF385A
|
zinc finger protein 385A
|
|
chr6_+_31651319
|
1.180
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr6_-_88911708
|
1.176
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr19_-_6432765
|
1.169
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr19_-_17349096
|
1.167
|
NM_031310
|
PLVAP
|
plasmalemma vesicle associated protein
|
|
chr2_+_169637255
|
1.164
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr7_-_100077072
|
1.158
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr7_+_73826244
|
1.157
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr6_-_154719560
|
1.156
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr7_+_128367961
|
1.153
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr8_+_126511739
|
1.146
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr1_-_26517342
|
1.141
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chrX_-_135577102
|
1.135
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_-_169657395
|
1.135
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_+_156526182
|
1.133
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr11_-_64403712
|
1.132
|
|
EHD1
|
EH-domain containing 1
|
|
chrX_+_48427137
|
1.128
|
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr11_+_65164167
|
1.127
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr12_-_53661888
|
1.124
|
NM_001098815
|
KIAA0748
|
KIAA0748
|
|
chr11_+_2877486
|
1.120
|
NM_183233
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr7_-_132371208
|
1.115
|
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr6_-_31658044
|
1.109
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chrX_+_48427103
|
1.105
|
NM_000377
|
WAS
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia)
|
|
chr18_-_50004770
|
1.102
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr15_+_36332258
|
1.100
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr17_+_44642587
|
1.097
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr19_-_8548232
|
1.093
|
|
MYO1F
|
myosin IF
|
|
chr7_+_18296569
|
1.092
|
|
HDAC9
|
histone deacetylase 9
|
|
chrX_-_134883798
|
1.091
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr8_-_116749382
|
1.091
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr11_+_66927955
|
1.087
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr16_+_48865521
|
1.087
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr17_-_55824083
|
1.086
|
|
USP32
|
ubiquitin specific peptidase 32
|
|
chr10_+_17312654
|
1.079
|
|
VIM
|
vimentin
|
|
chr5_+_140871107
|
1.071
|
|
|
|
|
chr17_-_59363352
|
1.055
|
NM_000626
NM_001039933
NM_021602
|
CD79B
|
CD79b molecule, immunoglobulin-associated beta
|
|
chr19_+_40465249
|
1.050
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr22_+_23333610
|
1.050
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr3_-_15349086
|
1.043
|
NM_004844
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated)
|
|
chr7_-_3049872
|
1.043
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr1_-_180908639
|
1.036
|
NM_001102450
NM_033345
|
RGS8
|
regulator of G-protein signaling 8
|
|
chr7_+_149779011
|
1.033
|
|
GIMAP8
|
GTPase, IMAP family member 8
|
|
chr1_+_154351162
|
1.028
|
|
LMNA
|
lamin A/C
|
|
chr16_+_31274020
|
1.027
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr5_-_169657312
|
1.020
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_-_169657360
|
1.018
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr11_+_46323575
|
1.012
|
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr11_+_59902530
|
1.010
|
NM_021201
NM_206938
NM_206939
NM_206940
|
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7
|
|
chr7_+_106293112
|
1.010
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr3_-_122862447
|
1.009
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr22_+_23153529
|
1.007
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr19_+_54068373
|
0.997
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr11_+_118269310
|
0.996
|
NM_032966
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr11_+_46323495
|
0.993
|
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr6_+_144649142
|
0.991
|
|
UTRN
|
utrophin
|
|
chr20_-_1548589
|
0.987
|
NM_001083910
NM_001135844
NM_006065
|
SIRPB1
|
signal-regulatory protein beta 1
|
|
chr19_-_15436333
|
0.986
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr12_-_8656705
|
0.982
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr11_-_117640078
|
0.977
|
|
|
|
|
chr5_-_169657192
|
0.977
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr2_-_231498184
|
0.972
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr10_-_49483002
|
0.971
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr6_+_108988797
|
0.970
|
|
FOXO3
|
forkhead box O3
|
|
chr3_-_120861959
|
0.968
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr1_+_78126787
|
0.968
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr12_+_52103905
|
0.966
|
NM_001164690
NM_001164691
NM_020547
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr16_+_80086454
|
0.965
|
NM_030629
|
CMIP
|
c-Maf-inducing protein
|
|
chr1_+_36462583
|
0.964
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr3_-_122862391
|
0.960
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr1_+_51474558
|
0.958
|
|
RNF11
|
ring finger protein 11
|
|
chr19_-_56726704
|
0.952
|
NM_001177547
NM_001177548
NM_001177549
NM_001245
NM_198845
NM_198846
|
SIGLEC6
|
sialic acid binding Ig-like lectin 6
|
|
chr5_-_176869440
|
0.949
|
|
DOK3
|
docking protein 3
|
|
chr2_-_168812132
|
0.947
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr19_+_59865935
|
0.944
|
NM_001081438
NM_006847
|
LILRB4
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4
|
|
chr5_+_159589014
|
0.940
|
NM_001445
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr17_+_35532994
|
0.939
|
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr10_-_71002998
|
0.932
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr7_+_150065376
|
0.927
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr22_+_42908612
|
0.919
|
|
PARVG
|
parvin, gamma
|
|
chr6_+_37081387
|
0.917
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr1_-_27113018
|
0.915
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr16_+_31273969
|
0.912
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr7_-_81237162
|
0.910
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr1_+_111544469
|
0.908
|
|
|
|
|
chr11_-_76059435
|
0.908
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr5_-_157218555
|
0.908
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr1_-_158815895
|
0.907
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr1_+_156590109
|
0.901
|
NM_001042583
NM_001042584
NM_001042585
NM_001042586
NM_001042587
NM_001185107
NM_001185108
NM_001185110
NM_001185112
NM_001185113
NM_001185114
NM_001185115
NM_030893
|
CD1E
|
CD1e molecule
|
|
chr1_+_43539239
|
0.898
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr2_+_241213334
|
0.896
|
NM_005301
|
GPR35
|
G protein-coupled receptor 35
|
|
chr11_+_1818007
|
0.890
|
NM_001145841
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr3_-_187025501
|
0.889
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_-_205161821
|
0.889
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr10_+_99884370
|
0.888
|
NM_014472
|
C10orf28
|
chromosome 10 open reading frame 28
|
|
chr3_+_3056307
|
0.884
|
NM_175612
|
CNTN4
|
contactin 4
|
|
chr16_-_30301634
|
0.883
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr17_-_35975227
|
0.882
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr1_+_157441824
|
0.879
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr1_+_51474502
|
0.876
|
|
RNF11
|
ring finger protein 11
|