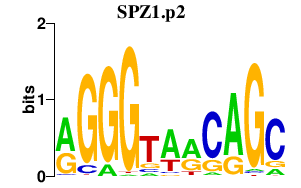
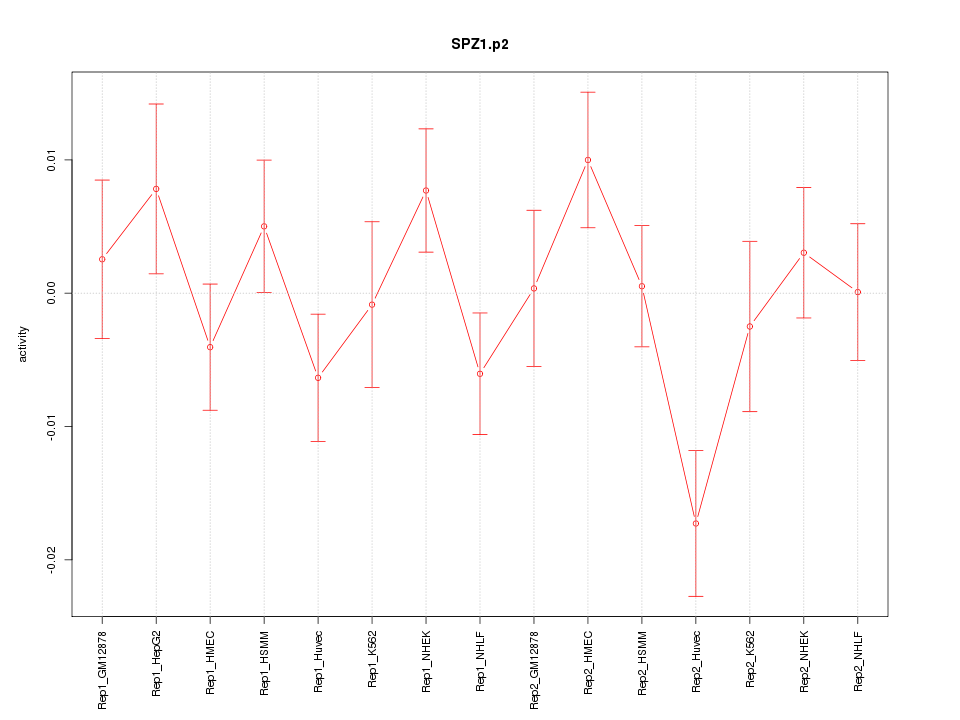
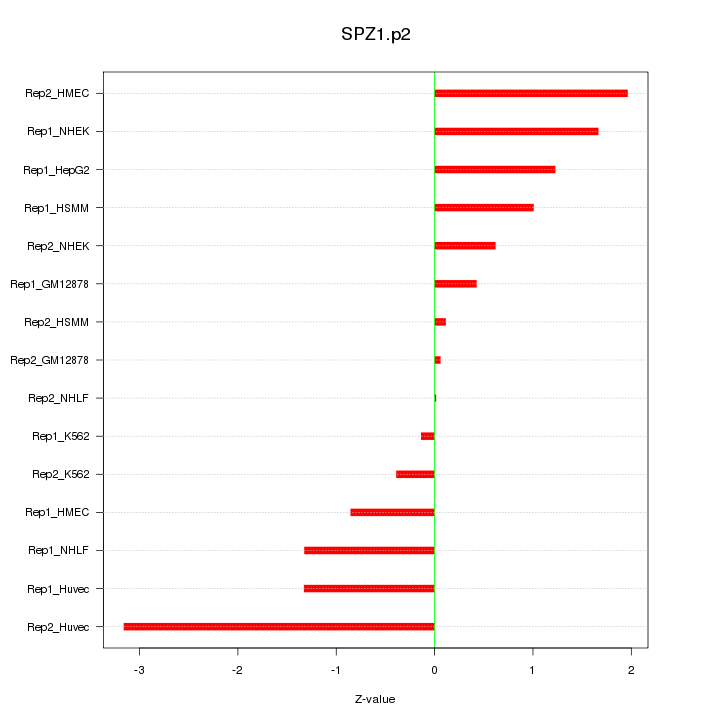
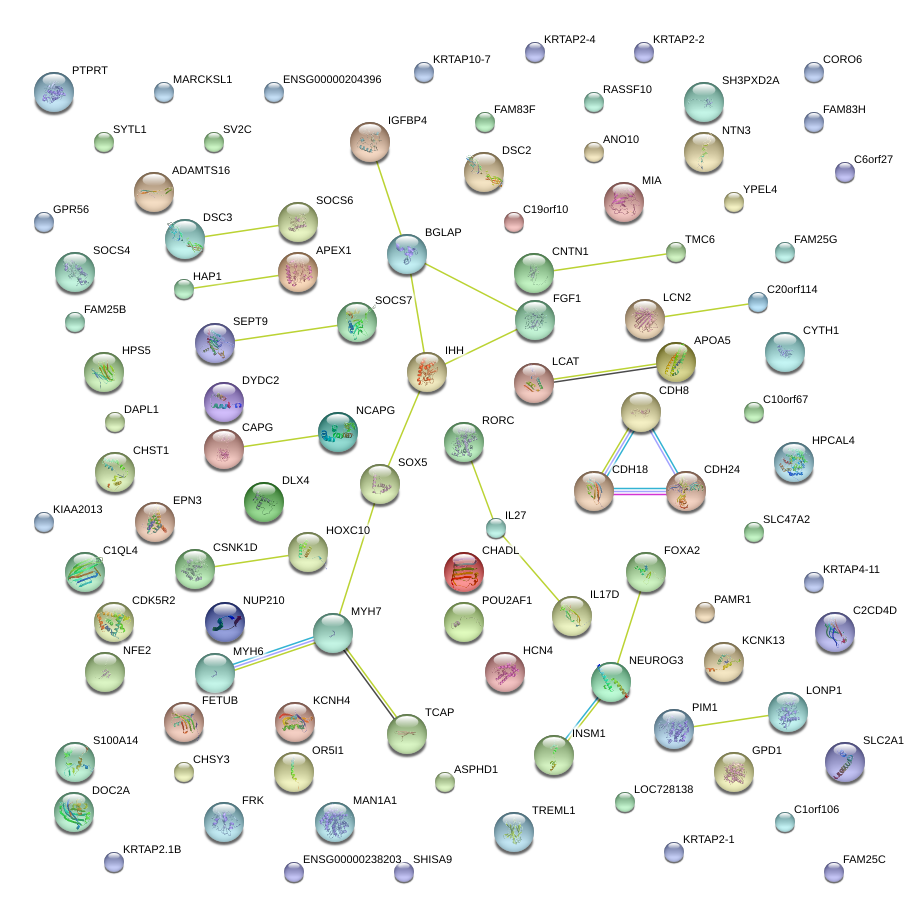
|
chr12_-_52980914
|
0.866
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr1_+_199130571
|
0.854
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr17_-_36469869
|
0.797
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr19_+_45973139
|
0.745
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr3_+_214668
|
0.738
|
|
|
|
|
chr16_+_29819647
|
0.724
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr10_-_105411339
|
0.716
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr17_-_74231266
|
0.673
|
|
CYTH1
|
cytohesin 1
|
|
chr17_-_24973940
|
0.662
|
|
CORO6
|
coronin 6
|
|
chr17_-_36475652
|
0.643
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr11_+_12987455
|
0.634
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr6_-_41230003
|
0.608
|
NM_178174
|
TREML1
|
triggering receptor expressed on myeloid cells-like 1
|
|
chr1_-_151855413
|
0.608
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr6_-_119712596
|
0.590
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr3_-_62835601
|
0.584
|
|
|
|
|
chr17_-_77824849
|
0.580
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr11_-_110755366
|
0.565
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr12_-_52980984
|
0.565
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr16_-_66535457
|
0.560
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr17_+_33761234
|
0.558
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr1_-_150070849
|
0.557
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr14_-_22974707
|
0.550
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr17_+_45405128
|
0.535
|
NM_001934
|
DLX4
|
distal-less homeobox 4
|
|
chr2_-_219633481
|
0.534
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr16_+_2461500
|
0.534
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr18_-_26935883
|
0.531
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_-_32574185
|
0.520
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr1_+_27541086
|
0.514
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr6_+_37245860
|
0.510
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr11_-_57171554
|
0.509
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr21_+_44844924
|
0.492
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr17_+_35075124
|
0.487
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr17_-_74231240
|
0.486
|
|
CYTH1
|
cytohesin 1
|
|
chr14_-_22596522
|
0.478
|
NM_022478
NM_144985
|
CDH24
|
cadherin 24, type 2
|
|
chr17_-_73640082
|
0.477
|
NM_007267
|
TMC6
|
transmembrane channel-like 6
|
|
chr16_-_28425632
|
0.475
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr11_-_116168316
|
0.464
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr17_+_45965044
|
0.461
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr10_+_47867667
|
0.452
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr17_-_36456949
|
0.451
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr10_-_23673777
|
0.449
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr1_-_11909042
|
0.449
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr10_+_82106537
|
0.440
|
NM_032372
|
DYDC2
|
DPY30 domain containing 2
|
|
chr17_-_37586716
|
0.438
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr11_-_35503720
|
0.435
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr12_+_48784057
|
0.430
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr1_-_150079656
|
0.429
|
NM_001136003
|
C2CD4D
|
C2 calcium-dependent domain containing 4D
|
|
chr2_-_85490984
|
0.425
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr16_+_56211150
|
0.424
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr8_+_142471290
|
0.424
|
|
|
|
|
chr11_-_45643711
|
0.422
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr3_+_187840842
|
0.421
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr5_-_142046175
|
0.414
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_+_154478574
|
0.414
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr15_-_71448194
|
0.410
|
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr17_+_72795567
|
0.409
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr16_+_56211410
|
0.406
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr10_-_46601687
|
0.405
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr10_-_71002998
|
0.405
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr2_+_219532590
|
0.405
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr20_-_22514092
|
0.402
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr5_+_129268352
|
0.402
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr15_-_71448657
|
0.399
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr1_-_43168247
|
0.398
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr9_+_129951552
|
0.396
|
NM_005564
|
LCN2
|
lipocalin 2
|
|
chr6_-_116488458
|
0.396
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr16_+_65579988
|
0.395
|
NM_173815
|
CES4A
|
carboxylesterase 4A
|
|
chr12_-_24606616
|
0.394
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr8_-_144887876
|
0.394
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr22_+_38720881
|
0.393
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr15_-_19221463
|
0.391
|
|
|
|
|
chr3_-_13436802
|
0.388
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr6_-_31853027
|
0.385
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr5_+_75414994
|
0.384
|
NM_014979
|
SV2C
|
synaptic vesicle glycoprotein 2C
|
|
chr1_-_39929654
|
0.383
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr2_-_85490997
|
0.382
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr20_+_31334601
|
0.381
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr12_+_52665196
|
0.380
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr12_+_39372456
|
0.379
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr12_-_48017223
|
0.379
|
NM_001008223
|
C1QL4
|
complement component 1, q subcomponent-like 4
|
|
chr17_-_36528113
|
0.379
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr17_-_37144408
|
0.378
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr17_+_35853177
|
0.378
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr14_+_89597860
|
0.377
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr16_+_12902955
|
0.370
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr17_-_19560470
|
0.370
|
NM_001099646
NM_152908
|
SLC47A2
|
solute carrier family 47, member 2
|
|
chr16_-_29929801
|
0.369
|
NM_003586
|
DOC2A
|
double C2-like domains, alpha
|
|
chr16_-_60627536
|
0.369
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr5_+_5193442
|
0.368
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr22_-_39964001
|
0.364
|
|
CHADL
|
chondroadherin-like
|
|
chr2_+_159360074
|
0.364
|
NM_001017920
|
DAPL1
|
death associated protein-like 1
|
|
chr20_+_20296744
|
0.363
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr15_-_73792234
|
0.363
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr12_+_47658502
|
0.360
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr19_-_6671585
|
0.359
|
NM_000064
|
C3
|
complement component 3
|
|
chr16_+_22432344
|
0.359
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr20_+_60558369
|
0.358
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chrX_-_106033202
|
0.357
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr11_-_35397090
|
0.357
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr17_+_70431964
|
0.352
|
NM_178160
|
OTOP2
|
otopetrin 2
|
|
chr2_+_232951487
|
0.349
|
NM_001632
|
ALPP
|
alkaline phosphatase, placental
|
|
chr17_-_40858775
|
0.347
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr11_+_19691397
|
0.346
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chrX_+_105856531
|
0.346
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr17_-_40344203
|
0.345
|
|
GFAP
|
glial fibrillary acidic protein
|
|
chr19_+_12810258
|
0.344
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr11_+_67007155
|
0.343
|
|
AIP
|
aryl hydrocarbon receptor interacting protein
|
|
chr20_-_30636535
|
0.342
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr13_+_40929525
|
0.341
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr14_-_64416307
|
0.340
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr17_-_35017650
|
0.340
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr9_-_106705867
|
0.338
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_+_14797764
|
0.338
|
NM_015209
NM_201628
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr17_-_45901173
|
0.337
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr6_-_30151526
|
0.334
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr2_-_36384
|
0.333
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr15_-_81113676
|
0.333
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr17_+_41217435
|
0.332
|
|
CRHR1
|
corticotropin releasing hormone receptor 1
|
|
chr22_-_44751671
|
0.332
|
NM_058238
|
WNT7B
|
wingless-type MMTV integration site family, member 7B
|
|
chr15_+_38437703
|
0.331
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr14_+_22581215
|
0.330
|
NM_001099780
|
PSMB11
|
proteasome (prosome, macropain) subunit, beta type, 11
|
|
chr16_+_29373365
|
0.330
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr10_-_118419459
|
0.330
|
NM_144661
|
C10orf82
|
chromosome 10 open reading frame 82
|
|
chr1_-_41722883
|
0.330
|
NM_001956
|
EDN2
|
endothelin 2
|
|
chr2_-_127580975
|
0.329
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr17_+_71582486
|
0.329
|
NM_003857
|
GALR2
|
galanin receptor 2
|
|
chr11_+_67007080
|
0.329
|
NM_003977
|
AIP
|
aryl hydrocarbon receptor interacting protein
|
|
chr20_-_30535045
|
0.327
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr19_+_53747233
|
0.326
|
NM_177973
|
SULT2B1
|
sulfotransferase family, cytosolic, 2B, member 1
|
|
chr5_-_134899537
|
0.325
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr6_-_31789824
|
0.325
|
|
|
|
|
chr3_-_13089547
|
0.324
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_+_151917674
|
0.320
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr7_-_156495985
|
0.319
|
NM_005515
|
MNX1
|
motor neuron and pancreas homeobox 1
|
|
chr1_+_176748834
|
0.318
|
NM_001170722
NM_001170723
NM_001170724
NM_032126
|
C1orf49
|
chromosome 1 open reading frame 49
|
|
chr16_-_66535923
|
0.317
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr21_+_44818033
|
0.317
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr16_+_658081
|
0.316
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr14_-_24588916
|
0.315
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr6_-_109882116
|
0.314
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr9_+_138863673
|
0.311
|
|
PHPT1
|
phosphohistidine phosphatase 1
|
|
chr1_+_3378030
|
0.309
|
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr22_-_49063088
|
0.309
|
|
PLXNB2
|
plexin B2
|
|
chr10_-_105410840
|
0.309
|
|
|
|
|
chr13_+_27264779
|
0.308
|
NM_145657
|
GSX1
|
GS homeobox 1
|
|
chr17_+_45993338
|
0.307
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr9_-_121171400
|
0.307
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chr12_+_55018925
|
0.307
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr20_-_34707971
|
0.306
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr18_-_26996742
|
0.306
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr18_-_55091596
|
0.304
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr2_-_231498184
|
0.304
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr8_+_35212505
|
0.304
|
NM_080872
|
UNC5D
|
unc-5 homolog D (C. elegans)
|
|
chr3_+_46898673
|
0.304
|
NM_001184744
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr8_-_22145462
|
0.304
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr22_-_43040063
|
0.303
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr3_+_125786195
|
0.302
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr1_-_1525309
|
0.302
|
|
LOC643988
|
hypothetical LOC643988
|
|
chr16_+_2954217
|
0.301
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr7_-_102070473
|
0.297
|
NM_001114403
|
UPK3BL
POLR2J2
|
uroplakin 3B-like
polymerase (RNA) II (DNA directed) polypeptide J2
|
|
chr2_+_241213334
|
0.296
|
NM_005301
|
GPR35
|
G protein-coupled receptor 35
|
|
chr1_+_46413335
|
0.295
|
NM_005727
|
TSPAN1
|
tetraspanin 1
|
|
chr10_-_71753684
|
0.293
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr20_-_33343617
|
0.293
|
NM_178468
|
FAM83C
|
family with sequence similarity 83, member C
|
|
chr3_-_187562470
|
0.292
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr15_-_73792150
|
0.292
|
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr4_-_120768563
|
0.289
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr16_-_87378872
|
0.285
|
NM_001142864
|
FAM38A
|
family with sequence similarity 38, member A
|
|
chr10_-_79298876
|
0.283
|
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr1_+_15935395
|
0.282
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr1_+_27541049
|
0.281
|
NM_001193308
NM_032872
|
SYTL1
|
synaptotagmin-like 1
|
|
chr11_+_27019084
|
0.280
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr16_-_4778317
|
0.279
|
NM_001154458
NM_144605
|
SEPT12
|
septin 12
|
|
chr21_+_26029542
|
0.279
|
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr17_-_40344322
|
0.277
|
|
|
|
|
chr17_-_35017691
|
0.277
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr8_+_65656164
|
0.277
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr13_-_71338558
|
0.276
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr4_+_995395
|
0.276
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr11_-_57039721
|
0.276
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr9_-_106705780
|
0.275
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr15_-_81412354
|
0.275
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr1_+_205137410
|
0.272
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr17_+_4348877
|
0.271
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr6_-_169392771
|
0.271
|
|
|
|
|
chr1_+_13782838
|
0.268
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr5_-_111782637
|
0.267
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr15_-_81037554
|
0.267
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr5_-_92942680
|
0.267
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr2_+_242400702
|
0.267
|
NM_001167599
NM_001167602
NM_080741
|
NEU4
|
sialidase 4
|
|
chr5_+_79021401
|
0.267
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr9_+_139241838
|
0.265
|
NM_001190228
|
LOC643596
|
hypothetical LOC643596
|
|
chr17_-_37034288
|
0.264
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr16_+_65765256
|
0.264
|
NM_003946
NM_001185057
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr17_-_74432671
|
0.264
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr15_+_97462675
|
0.263
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr14_+_20537253
|
0.263
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr18_-_22383066
|
0.263
|
NM_001136205
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr20_+_773283
|
0.262
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr1_+_203064397
|
0.262
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|