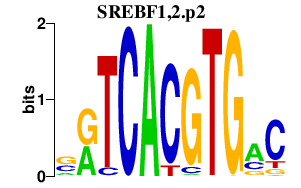
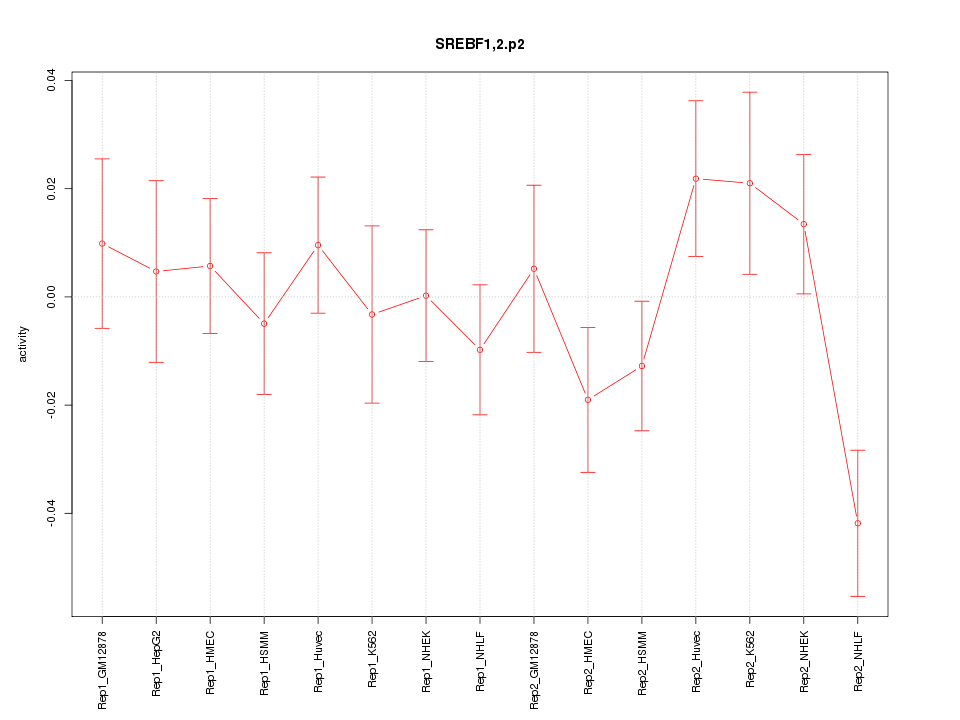
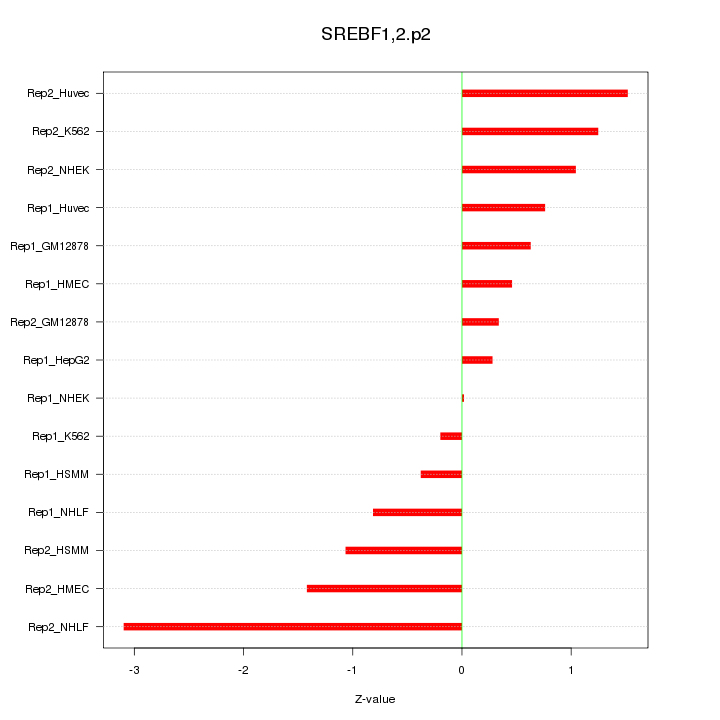
|
chr17_+_73638430
|
2.715
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr13_+_48582389
|
2.212
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr12_-_56432385
|
2.033
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr6_-_84994032
|
1.823
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr12_-_56432375
|
1.728
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432377
|
1.709
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr10_+_69314347
|
1.662
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr12_+_102982365
|
1.532
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr8_-_54918099
|
1.496
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chrX_+_101862297
|
1.486
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr19_+_45546369
|
1.465
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr6_+_160310118
|
1.347
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr5_+_133735004
|
1.321
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr10_-_45410197
|
1.315
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr11_+_125586828
|
1.276
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr8_-_54918389
|
1.239
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr17_+_69781980
|
1.221
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr11_-_125586733
|
1.218
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr2_-_172675873
|
1.202
|
|
DLX2
|
distal-less homeobox 2
|
|
chr19_+_37528340
|
1.124
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr16_-_28411123
|
1.109
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr1_-_26266571
|
1.105
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr20_+_43953606
|
1.079
|
|
CTSA
|
cathepsin A
|
|
chr12_-_56432292
|
1.077
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_27210330
|
1.069
|
NM_018405
|
C17orf79
|
chromosome 17 open reading frame 79
|
|
chr8_-_54918369
|
1.069
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr20_-_4743768
|
1.058
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr8_-_54918325
|
1.043
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr11_+_62295412
|
1.029
|
NM_006473
|
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa
|
|
chr1_-_39097904
|
0.995
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr11_-_85457753
|
0.983
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_+_20385164
|
0.973
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr16_+_1341937
|
0.970
|
|
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit
|
|
chr7_-_42918013
|
0.954
|
NM_001099858
NM_024054
|
C7orf25
|
chromosome 7 open reading frame 25
|
|
chr1_-_152797820
|
0.946
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr17_-_17816425
|
0.932
|
NM_001033551
NM_001082968
|
TOM1L2
|
target of myb1-like 2 (chicken)
|
|
chr2_+_219751230
|
0.932
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr3_-_20202622
|
0.931
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr10_-_50066386
|
0.930
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr12_+_67426184
|
0.927
|
NM_018656
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr6_+_87921980
|
0.921
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr2_-_20115269
|
0.910
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr3_-_123995216
|
0.899
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr19_-_17236522
|
0.898
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr10_+_112317401
|
0.877
|
NM_005445
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr7_-_44496703
|
0.860
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr5_+_41940216
|
0.857
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr20_+_37024383
|
0.857
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr13_-_49057707
|
0.855
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr10_+_25345513
|
0.827
|
NM_024838
|
THNSL1
|
threonine synthase-like 1 (S. cerevisiae)
|
|
chr16_+_22124870
|
0.822
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr10_+_69314714
|
0.819
|
|
SIRT1
|
sirtuin 1
|
|
chr13_-_49057629
|
0.818
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr3_-_121550767
|
0.816
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr17_+_63252704
|
0.803
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr2_+_227898163
|
0.802
|
|
MFF
|
mitochondrial fission factor
|
|
chr8_+_17824631
|
0.777
|
NM_006197
|
PCM1
|
pericentriolar material 1
|
|
chr5_+_34692124
|
0.773
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr12_+_88627601
|
0.767
|
|
LOC338758
|
hypothetical LOC338758
|
|
chr15_+_73415426
|
0.762
|
NM_017828
|
COMMD4
|
COMM domain containing 4
|
|
chr15_+_41871809
|
0.762
|
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 read-through transcript
small EDRK-rich factor 2
|
|
chr18_-_45241014
|
0.754
|
NM_017653
|
DYM
|
dymeclin
|
|
chr11_+_18300689
|
0.754
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr17_-_39556535
|
0.752
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr4_-_47160403
|
0.746
|
NM_017845
|
COMMD8
|
COMM domain containing 8
|
|
chr12_-_63439456
|
0.743
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_-_63439449
|
0.743
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr13_-_32757835
|
0.742
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_+_98725505
|
0.738
|
NM_178812
|
MTDH
|
metadherin
|
|
chr1_-_152797707
|
0.731
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr19_-_15436333
|
0.722
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr19_-_45546250
|
0.721
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr19_-_45546123
|
0.718
|
NM_178830
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr1_+_109221271
|
0.711
|
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr9_-_37894098
|
0.707
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr1_-_159459999
|
0.704
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr3_-_4483952
|
0.701
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr16_-_87570716
|
0.697
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr15_+_41871768
|
0.695
|
NM_001018108
|
SERF2
|
small EDRK-rich factor 2
|
|
chr1_+_181871830
|
0.695
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr11_-_85457736
|
0.690
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_-_152797767
|
0.687
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_+_123996655
|
0.686
|
|
DIRC2
|
disrupted in renal carcinoma 2
|
|
chr2_+_219751207
|
0.681
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chrX_-_85189194
|
0.678
|
NM_000390
NM_001145414
|
CHM
|
choroideremia (Rab escort protein 1)
|
|
chr12_-_54409107
|
0.676
|
NM_001040034
NM_001780
|
CD63
|
CD63 molecule
|
|
chr1_-_152797760
|
0.676
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr1_-_32054164
|
0.676
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr17_-_73636386
|
0.673
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr4_+_128773529
|
0.672
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr18_-_19271814
|
0.670
|
NM_032933
|
C18orf45
|
chromosome 18 open reading frame 45
|
|
chr20_+_43028533
|
0.665
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr3_-_4483955
|
0.659
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr4_-_47160476
|
0.657
|
|
COMMD8
|
COMM domain containing 8
|
|
chr8_-_74822206
|
0.656
|
|
STAU2
|
staufen, RNA binding protein, homolog 2 (Drosophila)
|
|
chr6_-_7258351
|
0.653
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr12_-_63439415
|
0.651
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_+_109963982
|
0.639
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr4_-_90197140
|
0.636
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr8_+_26296439
|
0.632
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr18_+_75967902
|
0.626
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr1_+_109963950
|
0.625
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chrX_-_100549444
|
0.622
|
|
GLA
|
galactosidase, alpha
|
|
chr3_-_4483939
|
0.619
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr6_-_44333002
|
0.618
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr2_+_208284484
|
0.617
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr1_-_165112157
|
0.617
|
NM_053053
|
TADA1
|
transcriptional adaptor 1
|
|
chr14_+_73621408
|
0.616
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr17_-_39556483
|
0.612
|
|
HDAC5
|
histone deacetylase 5
|
|
chr1_-_152867070
|
0.608
|
NM_001025107
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr20_-_39199936
|
0.606
|
|
|
|
|
chr19_+_10626106
|
0.605
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr4_+_56414565
|
0.605
|
NM_001024924
NM_018261
NM_178237
|
EXOC1
|
exocyst complex component 1
|
|
chr1_+_198263352
|
0.603
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr22_-_37966852
|
0.602
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog)
|
|
chr5_+_150807380
|
0.601
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr1_-_152867007
|
0.598
|
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr17_-_35510431
|
0.597
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr22_-_35233032
|
0.596
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr12_-_121316907
|
0.595
|
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr5_+_150807397
|
0.595
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr5_+_34692239
|
0.591
|
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_176575171
|
0.589
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr2_+_219751192
|
0.586
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr2_-_172572993
|
0.586
|
|
|
|
|
chr5_+_133734768
|
0.585
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr16_-_28410580
|
0.581
|
NM_000086
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr1_+_15725894
|
0.581
|
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr19_+_45546171
|
0.581
|
NM_001031696
NM_012268
|
PLD3
|
phospholipase D family, member 3
|
|
chr14_-_67353046
|
0.575
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr5_+_34692352
|
0.573
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr15_-_99609648
|
0.570
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr9_+_130749812
|
0.558
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr10_-_3817418
|
0.557
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr12_+_56271181
|
0.556
|
NM_001146258
NM_001146259
NM_001146260
NM_024779
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr11_-_6597154
|
0.555
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr11_-_18300246
|
0.554
|
NM_007216
NM_181507
NM_181508
|
HPS5
|
Hermansky-Pudlak syndrome 5
|
|
chr6_+_151815298
|
0.554
|
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr1_-_27099515
|
0.552
|
|
GPATCH3
|
G patch domain containing 3
|
|
chr2_+_169021001
|
0.550
|
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr10_-_112054497
|
0.549
|
|
SMNDC1
|
survival motor neuron domain containing 1
|
|
chr11_-_6597220
|
0.546
|
|
TPP1
|
tripeptidyl peptidase I
|
|
chr17_-_33043514
|
0.544
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr4_+_76658677
|
0.541
|
NM_144721
|
THAP6
|
THAP domain containing 6
|
|
chr2_+_208284610
|
0.535
|
|
CCNYL1
|
cyclin Y-like 1
|
|
chr21_-_35343065
|
0.535
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_+_17816851
|
0.531
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr16_-_1464927
|
0.530
|
|
CLCN7
|
chloride channel 7
|
|
chr6_-_163754903
|
0.530
|
|
LOC100526820
|
hypothetical LOC100526820
|
|
chr6_+_42126228
|
0.530
|
NM_138572
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr1_+_20081510
|
0.527
|
|
|
|
|
chr1_-_52936573
|
0.525
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr12_+_102883687
|
0.524
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr8_-_81246215
|
0.523
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr21_-_37561463
|
0.520
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr5_+_150807331
|
0.519
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr4_+_53975597
|
0.514
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr1_-_77920914
|
0.512
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr7_+_92699588
|
0.505
|
NM_017667
NM_024553
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr12_+_67426225
|
0.505
|
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr3_-_188945921
|
0.504
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_-_9908014
|
0.500
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr1_-_27099524
|
0.499
|
NM_022078
|
GPATCH3
|
G patch domain containing 3
|
|
chr21_+_29593098
|
0.498
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr5_+_139924595
|
0.498
|
NM_080670
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr1_-_40335451
|
0.496
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr17_-_35510074
|
0.493
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr16_+_5023672
|
0.492
|
|
LOC100507589
|
hypothetical LOC100507589
|
|
chr12_-_63439322
|
0.492
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_-_54409710
|
0.491
|
|
CD63
|
CD63 molecule
|
|
chr16_-_69876262
|
0.489
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr20_+_43028590
|
0.484
|
|
STK4
|
serine/threonine kinase 4
|
|
chr7_+_101860958
|
0.484
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_-_44037332
|
0.483
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr20_+_33823318
|
0.481
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr11_+_18300370
|
0.478
|
NM_001142307
NM_005316
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr19_+_7493436
|
0.478
|
NM_020533
|
MCOLN1
|
mucolipin 1
|
|
chr12_-_54409059
|
0.477
|
|
CD63
|
CD63 molecule
|
|
chr8_+_104102477
|
0.477
|
|
ATP6V1C1
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1
|
|
chr14_+_92869317
|
0.476
|
NM_020818
|
KIAA1409
|
KIAA1409
|
|
chr10_+_12211586
|
0.475
|
NM_001142627
NM_001142628
NM_018144
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr11_-_36267510
|
0.474
|
|
COMMD9
|
COMM domain containing 9
|
|
chr14_-_19999324
|
0.474
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chr5_+_149320630
|
0.472
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr11_-_85457469
|
0.472
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_61614930
|
0.472
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr10_+_70553862
|
0.470
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr5_-_122786902
|
0.466
|
NM_001166226
NM_153223
|
CEP120
|
centrosomal protein 120kDa
|
|
chr12_+_70435176
|
0.465
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr1_-_224253678
|
0.462
|
NM_152608
|
C1orf55
|
chromosome 1 open reading frame 55
|
|
chr1_-_239587007
|
0.462
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr1_+_26609819
|
0.462
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr6_+_42126242
|
0.461
|
|
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa
|
|
chr12_-_54409015
|
0.460
|
|
CD63
|
CD63 molecule
|
|
chr7_-_38915306
|
0.459
|
NM_014396
NM_080631
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae)
|
|
chr2_+_176689737
|
0.456
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr11_+_107385195
|
0.452
|
|
CUL5
|
cullin 5
|
|
chr14_-_23981493
|
0.452
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr2_+_172573049
|
0.451
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr22_+_23153529
|
0.451
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|