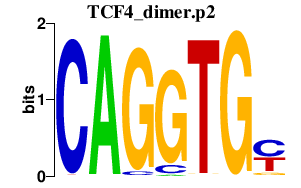
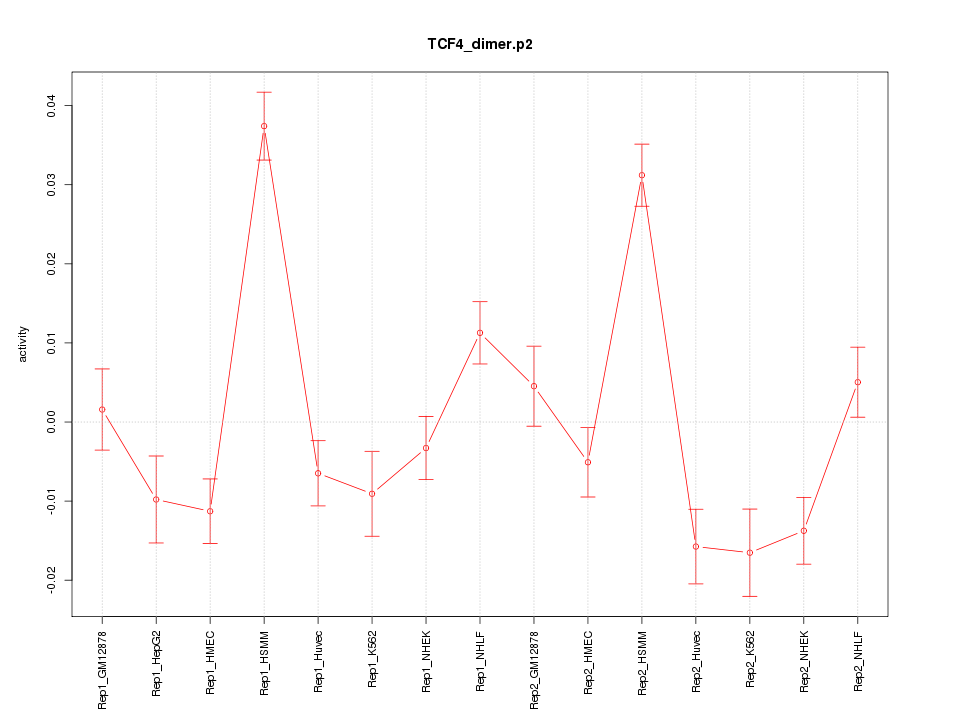
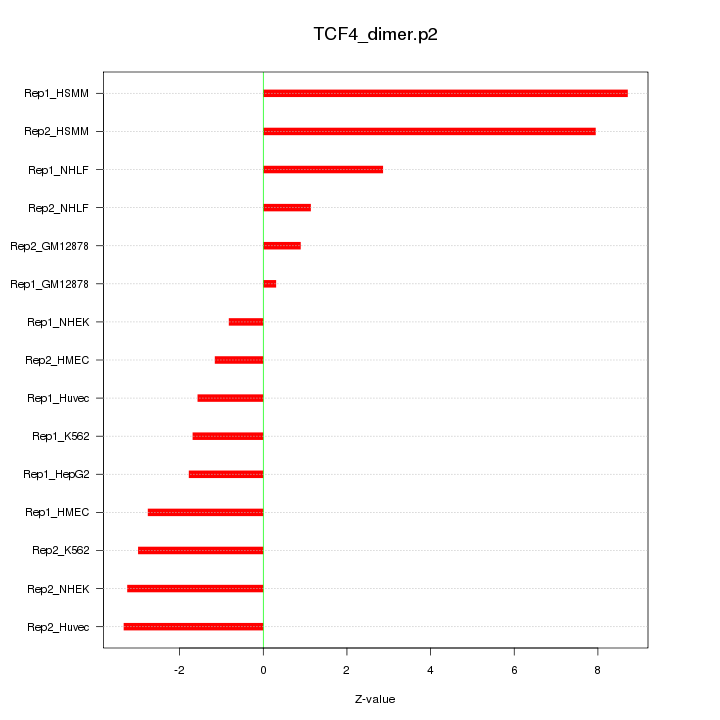
|
chr8_-_41641881
|
8.061
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr9_+_135389698
|
4.885
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr1_+_46151845
|
4.699
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr4_+_114190233
|
4.320
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_39209075
|
3.947
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr17_-_36451189
|
3.891
|
NM_030967
|
KRTAP1-3
KRTAP1-1
|
keratin associated protein 1-3
keratin associated protein 1-1
|
|
chr1_-_154205675
|
3.887
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr11_-_47427263
|
3.753
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr20_+_29870836
|
3.721
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr22_+_21094094
|
3.608
|
|
|
|
|
chr1_-_16217871
|
3.588
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr12_-_47679176
|
3.586
|
NM_015086
|
DDN
|
dendrin
|
|
chr2_+_219991342
|
3.558
|
NM_001927
|
DES
|
desmin
|
|
chr2_+_233099113
|
3.515
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr1_-_39871226
|
3.499
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr11_+_384238
|
3.452
|
|
PKP3
|
plakophilin 3
|
|
chr3_-_52844274
|
3.357
|
NM_205853
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr22_-_35011703
|
3.185
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr4_+_54660954
|
3.171
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr17_+_36493927
|
3.167
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr16_-_30983909
|
3.103
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr4_-_177427252
|
3.103
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr1_+_109969571
|
3.083
|
NM_203404
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr17_-_45629401
|
3.020
|
|
|
|
|
chr2_+_220007943
|
2.968
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr2_-_219826854
|
2.944
|
NM_006000
|
TUBA4A
|
tubulin, alpha 4a
|
|
chr2_+_27358800
|
2.877
|
NM_032546
NM_187841
|
TRIM54
|
tripartite motif containing 54
|
|
chr2_+_223625105
|
2.863
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr1_+_242283906
|
2.844
|
|
|
|
|
chr6_+_37245860
|
2.823
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_-_43168247
|
2.817
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr12_-_107745455
|
2.796
|
NM_001161331
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr9_+_139292094
|
2.795
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr7_+_1546535
|
2.787
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr11_+_384199
|
2.755
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr9_-_135379888
|
2.724
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr2_+_85515428
|
2.708
|
NM_198482
|
SH2D6
|
SH2 domain containing 6
|
|
chr1_+_201364050
|
2.707
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr11_+_63876924
|
2.681
|
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr15_+_61676604
|
2.677
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr15_-_49184764
|
2.660
|
NM_207381
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr11_-_62213946
|
2.657
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr8_-_93099040
|
2.657
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_201411533
|
2.650
|
NM_004997
|
MYBPH
|
myosin binding protein H
|
|
chr2_+_238712101
|
2.647
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr13_+_30378311
|
2.618
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr16_-_73798539
|
2.609
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr19_-_60740246
|
2.595
|
NM_001101401
|
SBK2
|
SH3-binding domain kinase family, member 2
|
|
chr1_-_153107084
|
2.565
|
|
|
|
|
chr16_+_7322751
|
2.515
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr2_-_166358792
|
2.463
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr6_-_31188310
|
2.431
|
NM_014070
|
C6orf15
|
chromosome 6 open reading frame 15
|
|
chr2_-_241408296
|
2.393
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr18_-_70110089
|
2.367
|
NM_001190807
NM_001914
NM_148923
|
CYB5A
|
cytochrome b5 type A (microsomal)
|
|
chr11_+_126378507
|
2.363
|
|
NCRNA00288
|
non-protein coding RNA 288
|
|
chr17_+_42641712
|
2.348
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr6_-_46401174
|
2.323
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr2_+_219453487
|
2.321
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr15_+_72397934
|
2.317
|
NM_182791
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr20_-_62052922
|
2.309
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr11_-_46674233
|
2.295
|
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chrX_-_33267646
|
2.293
|
NM_000109
|
DMD
|
dystrophin
|
|
chr11_-_65395248
|
2.279
|
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr4_-_152155827
|
2.267
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr15_-_40537002
|
2.260
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr1_+_26220852
|
2.241
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr5_-_176869148
|
2.231
|
|
|
|
|
chr6_-_53638464
|
2.219
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr19_+_54996613
|
2.211
|
|
|
|
|
chr1_+_6537924
|
2.199
|
NM_138697
NM_177540
|
TAS1R1
|
taste receptor, type 1, member 1
|
|
chr17_-_39094427
|
2.188
|
NM_004527
NM_013999
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr2_-_179380301
|
2.174
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr17_-_39094787
|
2.163
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr2_-_175337386
|
2.158
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr8_-_75396016
|
2.151
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr3_+_46894239
|
2.145
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr11_+_122936145
|
2.119
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr3_-_120861959
|
2.064
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr17_-_7434113
|
2.063
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr17_+_30498948
|
2.055
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr5_+_150008030
|
2.049
|
|
SYNPO
|
synaptopodin
|
|
chr3_-_69254226
|
2.037
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr15_-_72981595
|
2.032
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr11_-_129803515
|
2.031
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr12_-_6313219
|
2.012
|
|
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A
|
|
chr17_+_76549658
|
2.009
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr9_+_35782405
|
2.005
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr2_+_233112680
|
2.002
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chr22_+_29819343
|
1.996
|
|
SMTN
|
smoothelin
|
|
chr17_-_36444626
|
1.996
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr17_-_40695216
|
1.983
|
NM_152343
|
C17orf46
|
chromosome 17 open reading frame 46
|
|
chr11_+_1816808
|
1.978
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr3_+_42702014
|
1.968
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr1_+_15935395
|
1.965
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr19_-_15252261
|
1.956
|
NM_014299
NM_058243
|
BRD4
|
bromodomain containing 4
|
|
chr2_+_202379442
|
1.952
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr5_+_143171918
|
1.932
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr3_+_8750485
|
1.920
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr11_-_1464386
|
1.919
|
NM_001172223
|
MOB2
|
Mps one binder kinase activator-like 2
|
|
chr16_+_68284252
|
1.910
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr5_+_159276317
|
1.908
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr6_-_46401363
|
1.900
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr10_-_105208634
|
1.900
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr19_-_53914768
|
1.896
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr18_-_72973750
|
1.886
|
NM_001025100
NM_001025101
|
MBP
|
myelin basic protein
|
|
chr19_+_43447078
|
1.871
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_143643324
|
1.859
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_-_12747263
|
1.844
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr9_+_17568952
|
1.841
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr11_-_605649
|
1.827
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr1_+_144273662
|
1.816
|
|
ANKRD35
|
ankyrin repeat domain 35
|
|
chr7_+_73145481
|
1.814
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr2_+_85834111
|
1.806
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr12_-_51887266
|
1.804
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr11_+_65536037
|
1.799
|
NM_001323
|
CST6
|
cystatin E/M
|
|
chr15_-_23659329
|
1.797
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr17_+_36515166
|
1.797
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr10_-_75080784
|
1.793
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr12_+_79634820
|
1.789
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr15_+_72253224
|
1.782
|
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr7_+_129807467
|
1.781
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr1_-_143643064
|
1.774
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_+_43447262
|
1.773
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_+_26220895
|
1.767
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr14_-_70345485
|
1.766
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr1_+_54786394
|
1.765
|
NM_015547
NM_147161
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr10_-_102978678
|
1.752
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr19_+_43446937
|
1.745
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr2_-_217267742
|
1.736
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_72431274
|
1.731
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_-_70971556
|
1.730
|
NM_001005405
|
KRTAP5-11
|
keratin associated protein 5-11
|
|
chr22_-_16127520
|
1.718
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr10_+_3141596
|
1.715
|
|
PFKP
|
phosphofructokinase, platelet
|
|
chr20_+_19818209
|
1.712
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_-_51887318
|
1.703
|
|
ITGB7
|
integrin, beta 7
|
|
chr17_-_10393664
|
1.702
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr5_+_5193442
|
1.698
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr20_-_29896981
|
1.693
|
NM_004118
|
FOXS1
|
forkhead box S1
|
|
chr11_+_122936246
|
1.691
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr2_+_219532590
|
1.674
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr14_+_104244998
|
1.673
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr9_+_130224950
|
1.672
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr8_-_134184491
|
1.660
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr15_+_72253940
|
1.660
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr12_-_55931172
|
1.655
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr17_-_77474597
|
1.650
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr11_-_6298284
|
1.649
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr5_-_158690047
|
1.646
|
NM_002187
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40)
|
|
chr3_-_48445719
|
1.643
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr11_+_76516957
|
1.639
|
NM_000260
NM_001127179
NM_001127180
|
MYO7A
|
myosin VIIA
|
|
chr5_-_176869440
|
1.637
|
|
DOK3
|
docking protein 3
|
|
chr8_-_37576515
|
1.631
|
|
LOC157860
|
hypothetical protein LOC157860
|
|
chr9_-_116920232
|
1.625
|
NM_002160
|
TNC
|
tenascin C
|
|
chr1_-_201321711
|
1.618
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr6_-_46091238
|
1.616
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr1_-_12599934
|
1.613
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr11_+_70915960
|
1.613
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr12_+_120840829
|
1.612
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr11_-_70641267
|
1.610
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr4_-_1185144
|
1.608
|
|
LOC100130872
|
hypothetical LOC100130872
|
|
chr20_+_48781457
|
1.598
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr2_-_166358951
|
1.595
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr1_-_199348316
|
1.592
|
NM_000069
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr5_-_176869463
|
1.592
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr15_-_70385709
|
1.587
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_+_159943385
|
1.585
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr14_+_99220349
|
1.576
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr1_-_11830328
|
1.571
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr17_-_36456949
|
1.565
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr9_+_34980715
|
1.564
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr9_-_14769530
|
1.563
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr16_-_27982277
|
1.563
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr20_-_3635774
|
1.557
|
NM_023068
|
SIGLEC1
|
sialic acid binding Ig-like lectin 1, sialoadhesin
|
|
chr4_+_157044296
|
1.553
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr10_-_116517945
|
1.553
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr14_-_50480857
|
1.545
|
NM_001163940
NM_002863
|
PYGL
|
phosphorylase, glycogen, liver
|
|
chr6_-_33822659
|
1.543
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr13_-_32678142
|
1.542
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_-_166359016
|
1.541
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr1_+_238321794
|
1.538
|
NM_020066
|
FMN2
|
formin 2
|
|
chr2_-_152299229
|
1.534
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr1_+_226462363
|
1.532
|
NM_001098623
NM_052843
|
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr19_+_55627971
|
1.531
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr15_+_60838053
|
1.531
|
|
TLN2
|
talin 2
|
|
chr11_-_129803712
|
1.528
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr13_-_50315832
|
1.521
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr17_-_74487527
|
1.518
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr1_+_15145001
|
1.516
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_-_118739838
|
1.512
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr10_+_99322187
|
1.499
|
NM_001129981
NM_020349
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle)
|
|
chr3_+_120904555
|
1.499
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr22_+_16423147
|
1.499
|
NM_031481
|
SLC25A18
|
solute carrier family 25 (mitochondrial carrier), member 18
|
|
chr8_-_16903911
|
1.494
|
NM_019851
|
FGF20
|
fibroblast growth factor 20
|
|
chr7_-_127458231
|
1.494
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr7_-_100668820
|
1.492
|
NM_001185080
|
CLDN15
|
claudin 15
|
|
chr7_+_90176647
|
1.482
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr13_+_75232797
|
1.478
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr11_-_65841077
|
1.471
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr12_+_1799919
|
1.465
|
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr4_+_120276386
|
1.458
|
NM_016599
|
MYOZ2
|
myozenin 2
|