|
chr19_+_6415259
|
2.540
|
NM_139161
NM_174881
|
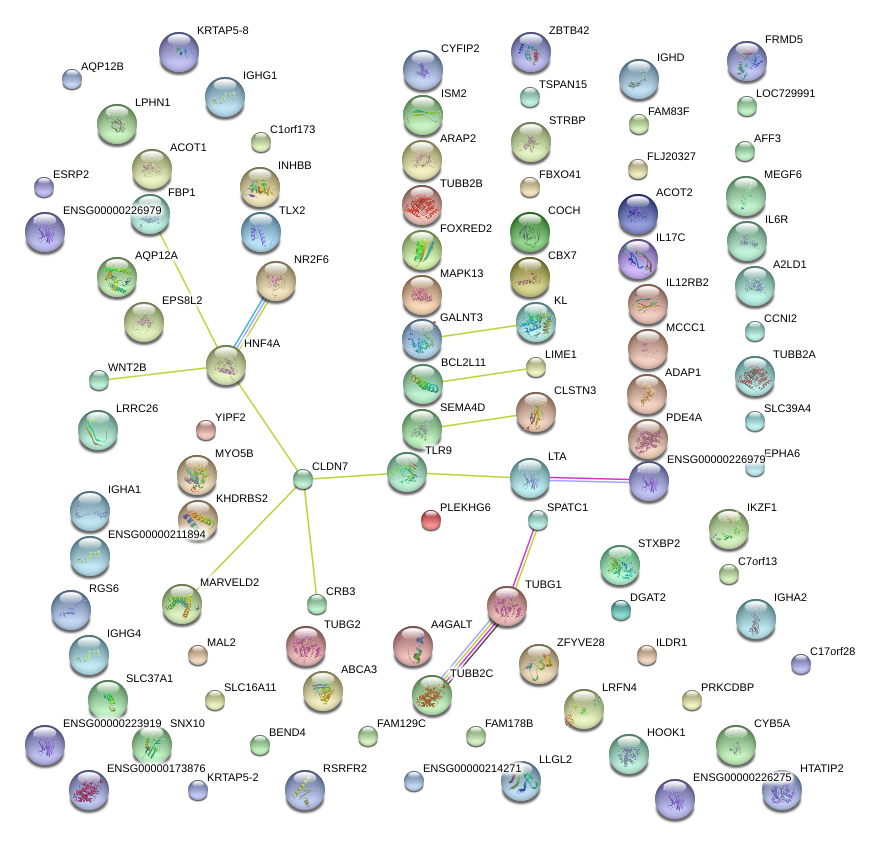
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr19_-_14177980
|
2.042
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr14_+_71468902
|
1.830
|
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr5_-_138753443
|
1.737
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr8_+_145158432
|
1.719
|
NM_001134374
NM_198572
|
SPATC1
|
spermatogenesis and centriole associated 1
|
|
chr17_-_60208083
|
1.622
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr2_-_97016027
|
1.582
|
NM_001122646
|
FAM178B
|
family with sequence similarity 178, member B
|
|
chr9_-_139183675
|
1.579
|
|
|
|
|
chr17_-_7106497
|
1.524
|
|
CLDN7
|
claudin 7
|
|
chr9_-_96441409
|
1.447
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr9_-_139184247
|
1.397
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr9_-_96441567
|
1.340
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr17_-_6887965
|
1.322
|
NM_153357
|
SLC16A11
|
solute carrier family 16, member 11 (monocarboxylic acid transporter 11)
|
|
chr14_+_30413393
|
1.200
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr5_-_157011665
|
1.161
|
|
|
|
|
chr3_-_184316553
|
1.159
|
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr22_-_41419800
|
1.152
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr9_-_96441654
|
1.137
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr6_+_36206239
|
1.116
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr5_+_68746694
|
1.102
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr17_+_38064787
|
1.072
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr15_-_42274760
|
1.041
|
|
FRMD5
|
FERM domain containing 5
|
|
chr1_+_1062137
|
1.002
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr1_+_112852892
|
0.999
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr17_-_7106987
|
0.986
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr3_-_123223658
|
0.983
|
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr8_+_120289735
|
0.977
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr21_+_42807052
|
0.974
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr22_+_38720881
|
0.974
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr1_-_3517918
|
0.969
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr7_-_927035
|
0.968
|
|
ADAP1
|
ArfGAP with dual PH domains 1
|
|
chr19_+_17499064
|
0.953
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr11_+_70926718
|
0.945
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr22_-_37858871
|
0.930
|
|
CBX7
|
chromobox homolog 7
|
|
chr8_-_145609737
|
0.922
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr19_-_6150536
|
0.913
|
|
|
|
|
chr2_+_120820140
|
0.912
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr2_-_241270982
|
0.906
|
NM_001102467
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr3_+_98016114
|
0.902
|
NM_001080448
|
EPHA6
|
EPH receptor A6
|
|
chr1_+_60053050
|
0.900
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr19_+_7608020
|
0.898
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr17_-_70480385
|
0.892
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr19_+_7608015
|
0.874
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr16_+_87232501
|
0.858
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr19_+_7607976
|
0.852
|
NM_001127396
NM_006949
|
STXBP2
|
syntaxin binding protein 2
|
|
chr11_+_75157414
|
0.851
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr19_-_17217022
|
0.849
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr19_+_10392128
|
0.841
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr9_-_125070675
|
0.836
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr11_+_75157450
|
0.821
|
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr4_-_35922288
|
0.815
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr19_-_7607852
|
0.815
|
|
|
|
|
chr14_-_105392671
|
0.801
|
|
|
|
|
chr2_+_74595073
|
0.787
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr6_-_63053811
|
0.786
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr16_-_66827449
|
0.777
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr7_+_26298136
|
0.777
|
|
SNX10
|
sorting nexin 10
|
|
chr2_+_111594921
|
0.775
|
NM_006538
NM_138621
NM_207002
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr19_+_7608010
|
0.775
|
|
STXBP2
|
syntaxin binding protein 2
|
|
chr13_-_100039008
|
0.773
|
NM_001195087
|
A2LD1
|
AIG2-like domain 1
|
|
chr12_+_6290127
|
0.771
|
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr4_-_41848874
|
0.758
|
|
BEND4
|
BEN domain containing 4
|
|
chr2_-_100125402
|
0.753
|
NM_002285
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr7_+_26298018
|
0.753
|
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr2_-_73350103
|
0.744
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr14_-_77034853
|
0.744
|
NM_182509
NM_199296
|
ISM2
|
isthmin 2 homolog (zebrafish)
|
|
chr9_-_91302707
|
0.739
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr6_-_3172775
|
0.725
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B
|
|
chr13_+_32488477
|
0.723
|
NM_004795
|
KL
|
klotho
|
|
chr6_+_31648071
|
0.720
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr18_-_70110089
|
0.710
|
NM_001190807
NM_001914
NM_148923
|
CYB5A
|
cytochrome b5 type A (microsomal)
|
|
chr3_-_52235218
|
0.703
|
NM_017442
|
TLR9
|
toll-like receptor 9
|
|
chr11_-_6298284
|
0.701
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr1_+_152644471
|
0.701
|
|
IL6R
|
interleukin 6 receptor
|
|
chr11_+_20342262
|
0.701
|
NM_001098523
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr17_+_71033339
|
0.696
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr5_+_132111034
|
0.695
|
NM_001039780
|
CCNI2
|
cyclin I family, member 2
|
|
chr17_-_70480345
|
0.692
|
|
|
|
|
chr12_+_6290359
|
0.690
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr15_-_42274529
|
0.689
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr1_+_67545634
|
0.685
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr22_-_35232979
|
0.683
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr14_+_104338423
|
0.675
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr11_+_712282
|
0.674
|
|
EPS8L2
|
EPS8-like 2
|
|
chr1_-_74912009
|
0.667
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr19_-_19142053
|
0.664
|
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr2_-_166359016
|
0.660
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr12_+_7174230
|
0.659
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr4_-_2390050
|
0.658
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr5_+_156625629
|
0.656
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr19_-_19142090
|
0.655
|
NM_001145785
|
MEF2B
|
myocyte enhancer factor 2B
|
|
chr10_+_70881322
|
0.648
|
|
|
|
|
chr14_+_73073570
|
0.646
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr7_-_156125876
|
0.634
|
|
C7orf13
|
chromosome 7 open reading frame 13
|
|
chr7_+_50314801
|
0.632
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr14_-_105851580
|
0.632
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr18_-_45975163
|
0.630
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr11_+_66381451
|
0.630
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr20_+_42463279
|
0.628
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_-_10900086
|
0.626
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr10_+_70881221
|
0.625
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr3_-_120236118
|
0.619
|
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr19_+_868285
|
0.619
|
NM_032551
|
KISS1R
|
KISS1 receptor
|
|
chr19_-_18409788
|
0.611
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr17_-_58877276
|
0.610
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr2_+_10779191
|
0.608
|
NM_001039362
NM_144583
|
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2
|
|
chr19_+_40431122
|
0.606
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr17_-_8643163
|
0.605
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr1_+_152644292
|
0.602
|
NM_000565
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr12_+_53229625
|
0.601
|
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr2_-_166358951
|
0.597
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr2_+_222997456
|
0.594
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr3_-_120236355
|
0.594
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr17_+_26839143
|
0.593
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr1_-_27834210
|
0.593
|
NM_005248
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr12_-_7793335
|
0.588
|
NM_130441
NM_203503
|
CLEC4C
|
C-type lectin domain family 4, member C
|
|
chr11_+_1287553
|
0.588
|
|
LOC255512
|
hypothetical LOC255512
|
|
chr1_+_67546143
|
0.583
|
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr1_+_205736095
|
0.582
|
NM_000573
NM_000651
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr9_-_125070613
|
0.579
|
NM_018387
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr12_+_6926014
|
0.579
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr18_-_5620629
|
0.577
|
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chr19_+_590878
|
0.577
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chrX_+_69591639
|
0.576
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr1_+_12046020
|
0.572
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr19_+_18391423
|
0.567
|
|
|
|
|
chr11_-_684842
|
0.567
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr7_-_37454933
|
0.562
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr5_+_176492629
|
0.561
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr17_+_68672705
|
0.560
|
NM_001050
|
SSTR2
|
somatostatin receptor 2
|
|
chr16_+_1524162
|
0.560
|
NM_024600
|
TMEM204
|
transmembrane protein 204
|
|
chr17_+_71229191
|
0.558
|
|
ITGB4
|
integrin, beta 4
|
|
chr19_+_17843753
|
0.554
|
NM_000453
|
SLC5A5
|
solute carrier family 5 (sodium iodide symporter), member 5
|
|
chr10_-_105208634
|
0.551
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr1_+_109458093
|
0.549
|
NM_020775
|
KIAA1324
|
KIAA1324
|
|
chr11_+_45900731
|
0.549
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr1_-_1840564
|
0.549
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr12_+_119562737
|
0.547
|
NM_001033677
|
CABP1
|
calcium binding protein 1
|
|
chr19_+_18391211
|
0.547
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr2_-_96145614
|
0.545
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr12_-_92489652
|
0.544
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr15_+_81567327
|
0.538
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr5_-_1647617
|
0.535
|
|
SDHAP3
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3
|
|
chr21_+_33364316
|
0.533
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr7_-_72822471
|
0.528
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr1_-_159257486
|
0.525
|
|
F11R
|
F11 receptor
|
|
chr2_-_25390266
|
0.524
|
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr15_+_72315719
|
0.521
|
NM_025055
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr11_-_1287256
|
0.518
|
|
TOLLIP
|
toll interacting protein
|
|
chr17_+_71229151
|
0.518
|
|
ITGB4
|
integrin, beta 4
|
|
chr20_+_42007949
|
0.515
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr5_-_132976094
|
0.514
|
|
FSTL4
|
follistatin-like 4
|
|
chr1_+_63561632
|
0.512
|
|
FOXD3
|
forkhead box D3
|
|
chr1_-_159257567
|
0.512
|
|
F11R
|
F11 receptor
|
|
chr11_-_72057755
|
0.511
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr5_-_132976121
|
0.506
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr11_-_1287384
|
0.505
|
|
TOLLIP
|
toll interacting protein
|
|
chr16_+_29374479
|
0.505
|
NM_001017389
NM_003166
|
SULT1A4
SULT1A3
SLX1B
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr9_-_139047112
|
0.504
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chr3_+_47004457
|
0.504
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chr1_-_58815415
|
0.504
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr19_+_45811647
|
0.504
|
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr18_+_58143499
|
0.501
|
NM_003839
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator
|
|
chr22_-_30071565
|
0.501
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr20_+_60744231
|
0.499
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr15_-_50375143
|
0.499
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr1_+_52871603
|
0.498
|
NM_001042693
|
FAM159A
|
family with sequence similarity 159, member A
|
|
chr1_+_109458299
|
0.498
|
|
KIAA1324
|
KIAA1324
|
|
chr20_-_10602139
|
0.497
|
|
JAG1
|
jagged 1
|
|
chr12_+_131797455
|
0.494
|
NM_001170543
NM_001170544
NM_138575
|
PGAM5
|
phosphoglycerate mutase family member 5
|
|
chr18_-_42815965
|
0.493
|
NM_016427
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2)
|
|
chr6_+_44295219
|
0.493
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr2_+_109112428
|
0.491
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr20_+_42463350
|
0.489
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr11_-_34491878
|
0.489
|
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr16_-_626293
|
0.487
|
NM_001040160
NM_001040161
NM_001040162
NM_001040165
NM_032366
|
C16orf13
|
chromosome 16 open reading frame 13
|
|
chr11_-_1287416
|
0.484
|
|
TOLLIP
|
toll interacting protein
|
|
chr2_+_46378405
|
0.484
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr17_-_34084682
|
0.483
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr1_-_109458001
|
0.482
|
NM_001122961
|
C1orf194
|
chromosome 1 open reading frame 194
|
|
chr11_-_1287467
|
0.479
|
NM_019009
|
TOLLIP
|
toll interacting protein
|
|
chr8_+_95722448
|
0.478
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr17_+_32368611
|
0.477
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr1_+_27433593
|
0.476
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr8_-_141537035
|
0.475
|
NM_001160372
|
TRAPPC9
|
trafficking protein particle complex 9
|
|
chr5_-_1348103
|
0.474
|
NM_001193376
NM_198253
|
TERT
|
telomerase reverse transcriptase
|
|
chr5_+_140997171
|
0.473
|
|
RELL2
|
RELT-like 2
|
|
chr11_-_407324
|
0.472
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr22_-_22426551
|
0.471
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr10_-_97040661
|
0.468
|
NM_020992
|
PDLIM1
|
PDZ and LIM domain 1
|
|
chr2_-_25245062
|
0.468
|
NM_000939
NM_001035256
|
POMC
|
proopiomelanocortin
|
|
chr17_+_71229110
|
0.467
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr14_-_105549177
|
0.466
|
|
IGHV4-31
IGHD
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr3_-_56810869
|
0.466
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_-_3097070
|
0.461
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr7_-_22363036
|
0.461
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr2_-_6923215
|
0.458
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr2_-_230287442
|
0.456
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr17_-_34015631
|
0.455
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr11_+_66643311
|
0.455
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|