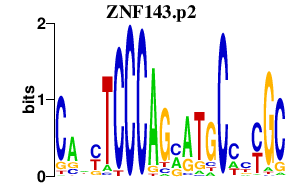
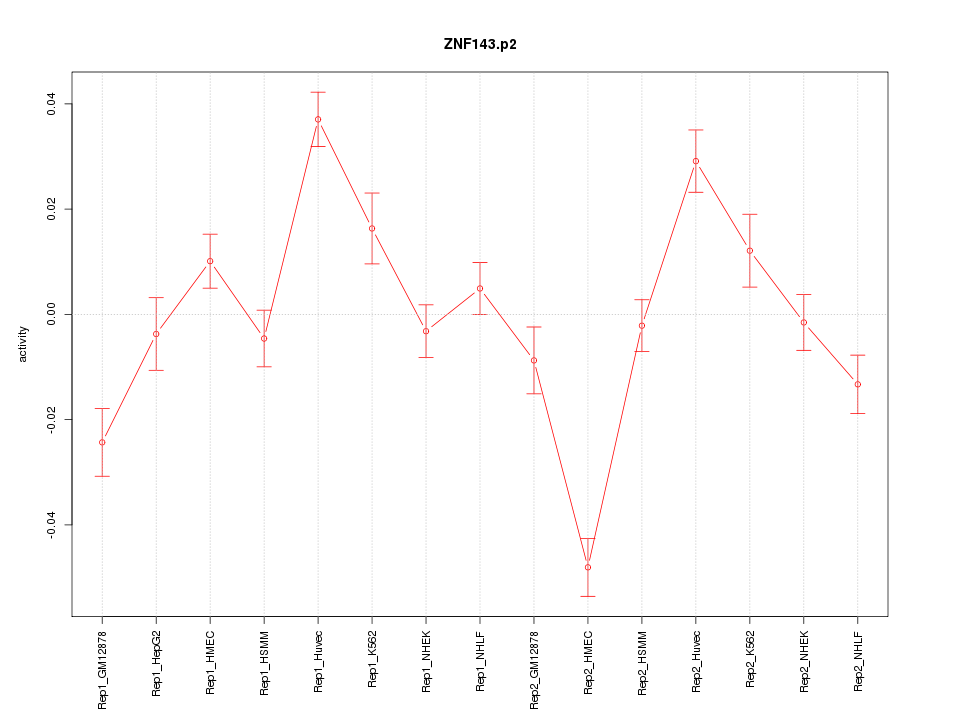
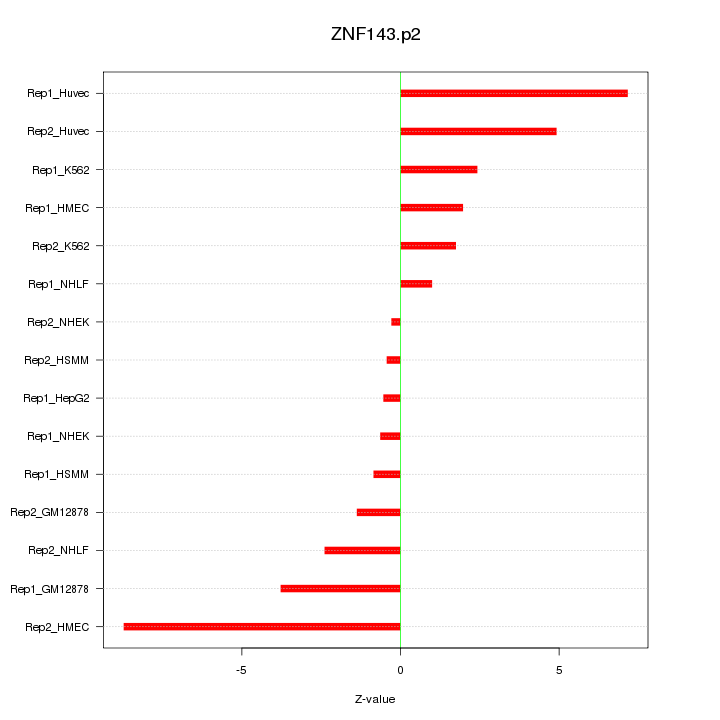
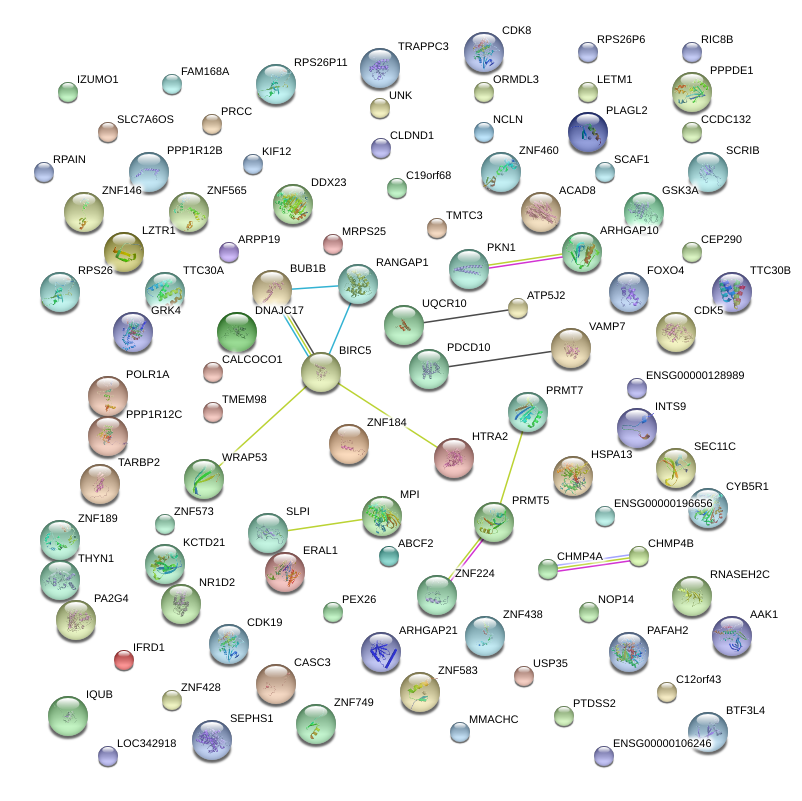
|
chr1_+_200584421
|
2.428
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr12_-_47532171
|
2.264
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr19_+_3136965
|
2.130
|
|
NCLN
|
nicalin
|
|
chr12_+_87060176
|
2.052
|
NM_181783
|
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3
|
|
chr7_-_150385912
|
1.952
|
NM_001164410
NM_004935
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr11_-_72986738
|
1.936
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr19_-_60320592
|
1.867
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory (inhibitor) subunit 12C
|
|
chr22_+_19666508
|
1.821
|
NM_006767
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr7_-_150385867
|
1.781
|
|
CDK5
|
cyclin-dependent kinase 5
|
|
chr7_-_150555153
|
1.746
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr15_-_38886961
|
1.675
|
NM_018163
|
DNAJC17
|
DnaJ (Hsp40) homolog, subfamily C, member 17
|
|
chr17_-_35337288
|
1.573
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr6_+_11202251
|
1.556
|
NM_001135575
|
LOC221710
|
hypothetical protein LOC221710
|
|
chr2_+_74610751
|
1.550
|
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr20_+_31862789
|
1.544
|
|
CHMP4B
|
chromatin modifying protein 4B
|
|
chr11_-_65244486
|
1.534
|
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr22_+_28493351
|
1.491
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr15_-_38886937
|
1.487
|
|
DNAJC17
|
DnaJ (Hsp40) homolog, subfamily C, member 17
|
|
chr1_+_45738442
|
1.464
|
NM_015506
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria
|
|
chr1_-_26197121
|
1.443
|
NM_000437
|
PAFAH2
|
platelet-activating factor acetylhydrolase 2, 40kDa
|
|
chr1_+_154291140
|
1.433
|
NM_001145264
NM_014017
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2
|
|
chr20_-_30259007
|
1.428
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr22_+_19666616
|
1.426
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr12_+_105692466
|
1.423
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr21_-_14677310
|
1.407
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr4_-_1827475
|
1.394
|
NM_012318
|
LETM1
|
leucine zipper-EF-hand containing transmembrane protein 1
|
|
chr12_+_54721952
|
1.394
|
NM_001029
|
RPS26
|
ribosomal protein S26
|
|
chr4_+_2935065
|
1.371
|
NM_001004056
NM_001004057
NM_182982
|
GRK4
|
G protein-coupled receptor kinase 4
|
|
chr2_-_69724480
|
1.350
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr22_+_16940623
|
1.346
|
NM_001127649
NM_017929
|
PEX26
|
peroxisomal biogenesis factor 26
|
|
chr3_-_168935287
|
1.332
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr22_-_40012115
|
1.325
|
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chr22_+_16940755
|
1.316
|
|
PEX26
|
peroxisomal biogenesis factor 26
|
|
chr10_-_13430195
|
1.270
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr12_+_52180971
|
1.268
|
NM_004178
NM_134323
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr19_+_53365748
|
1.268
|
|
C19orf68
|
chromosome 19 open reading frame 68
|
|
chr17_+_73721871
|
1.266
|
NM_001012270
NM_001012271
NM_001168
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr22_+_19666575
|
1.262
|
|
LZTR1
|
leucine-zipper-like transcription regulator 1
|
|
chr1_+_155003897
|
1.249
|
NM_005973
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr7_+_92699588
|
1.228
|
NM_017667
NM_024553
|
CCDC132
|
coiled-coil domain containing 132
|
|
chr19_+_62483606
|
1.219
|
NM_006635
|
ZNF460
|
zinc finger protein 460
|
|
chr19_-_42962039
|
1.207
|
NM_001172689
NM_001172690
NM_001172691
NM_001172692
NM_152360
|
ZNF573
|
zinc finger protein 573
|
|
chr7_+_111877699
|
1.200
|
NM_001550
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr8_-_28803616
|
1.194
|
NM_001145159
NM_018250
|
INTS9
|
integrator complex subunit 9
|
|
chr20_+_31862707
|
1.191
|
NM_176812
|
CHMP4B
|
chromatin modifying protein 4B
|
|
chr16_-_66902286
|
1.181
|
NM_032178
|
SLC7A6OS
|
solute carrier family 7, member 6 opposite strand
|
|
chr12_+_54784681
|
1.170
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr15_+_38240555
|
1.169
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr17_+_71292535
|
1.163
|
|
|
|
|
chr13_+_25726238
|
1.126
|
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr12_-_87059877
|
1.123
|
NM_025114
|
CEP290
|
centrosomal protein 290kDa
|
|
chr19_-_47438583
|
1.123
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr8_-_28803341
|
1.120
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr11_-_77577288
|
1.114
|
NM_001029859
|
KCTD21
|
potassium channel tetramerisation domain containing 21
|
|
chr12_+_52181643
|
1.113
|
NM_134324
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr1_+_155003939
|
1.111
|
|
PRCC
|
papillary renal cell carcinoma (translocation-associated)
|
|
chr19_-_53941835
|
1.107
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr15_+_72969404
|
1.104
|
NM_002435
|
MPI
|
mannose phosphate isomerase
|
|
chr1_-_201202956
|
1.103
|
NM_016243
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr19_+_61607505
|
1.099
|
NM_152478
NM_001159861
|
ZNF583
|
zinc finger protein 583
|
|
chr19_-_47438548
|
1.081
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr17_+_73721924
|
1.079
|
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr8_-_144969529
|
1.073
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr19_-_48815572
|
1.068
|
|
ZNF428
|
zinc finger protein 428
|
|
chr17_+_24206100
|
1.067
|
NM_005702
|
ERAL1
|
Era G-protein-like 1 (E. coli)
|
|
chr6_-_27548438
|
1.066
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr1_-_36387653
|
1.048
|
NM_014408
|
TRAPPC3
|
trafficking protein particle complex 3
|
|
chr3_-_99724047
|
1.041
|
|
CLDND1
|
claudin domain containing 1
|
|
chr17_+_71292437
|
1.026
|
|
UNK
|
unkempt homolog (Drosophila)
|
|
chr19_-_41397396
|
1.024
|
NM_152477
|
ZNF565
|
zinc finger protein 565
|
|
chr19_+_54837181
|
1.023
|
NM_021228
|
SCAF1
|
SR-related CTD-associated factor 1
|
|
chr15_-_50648589
|
1.022
|
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr17_+_71292275
|
1.005
|
NM_001080419
|
UNK
|
unkempt homolog (Drosophila)
|
|
chr12_-_52407470
|
1.002
|
NM_001143682
NM_020898
|
CALCOCO1
|
calcium binding and coiled-coil domain 1
|
|
chr16_+_66902369
|
0.999
|
NM_001184824
NM_019023
|
PRMT7
|
protein arginine methyltransferase 7
|
|
chr1_+_52294444
|
0.994
|
NM_001136497
NM_152265
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr17_+_35550292
|
0.991
|
|
CASC3
|
cancer susceptibility candidate 3
|
|
chr1_+_52294563
|
0.986
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr17_+_35550318
|
0.983
|
|
|
|
|
chr7_-_98901699
|
0.976
|
NM_001198879
NM_001190353
NM_001190354
NM_001003713
NM_001003714
NM_001039178
NM_004889
|
ATP5J2-PTCD1
ATP5J2
|
ATP5J2-PTCD1 readthrough
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2
|
|
chr10_-_13429397
|
0.962
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr10_-_31360778
|
0.961
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr16_-_219111
|
0.961
|
|
|
|
|
chr10_-_13430024
|
0.959
|
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr2_-_86186441
|
0.954
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr15_-_50648636
|
0.954
|
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr19_-_47438563
|
0.952
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr4_+_148872902
|
0.946
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr9_+_103200939
|
0.943
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr1_+_242883057
|
0.939
|
|
PPPDE1
|
PPPDE peptidase domain containing 1
|
|
chr11_+_440408
|
0.938
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr12_-_119938667
|
0.937
|
NM_022895
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr17_+_28279283
|
0.934
|
|
TMEM98
|
transmembrane protein 98
|
|
chr19_-_48815853
|
0.926
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr1_+_52294554
|
0.925
|
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr9_-_115901157
|
0.918
|
NM_138424
|
KIF12
|
kinesin family member 12
|
|
chr11_-_133628348
|
0.911
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr6_+_11201977
|
0.911
|
|
LOC221710
|
hypothetical protein LOC221710
|
|
chrX_+_154764136
|
0.902
|
NM_001145149
NM_001185183
NM_005638
|
VAMP7
|
vesicle-associated membrane protein 7
|
|
chr1_-_36387458
|
0.902
|
|
TRAPPC3
|
trafficking protein particle complex 3
|
|
chr18_+_54958095
|
0.898
|
NM_033280
|
SEC11C
|
SEC11 homolog C (S. cerevisiae)
|
|
chr12_+_54784632
|
0.897
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr19_-_41711009
|
0.892
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr19_+_3136860
|
0.891
|
NM_020170
|
NCLN
|
nicalin
|
|
chr19_+_41397343
|
0.891
|
NM_001099638
NM_001099639
NM_007145
|
ZNF146
|
zinc finger protein 146
|
|
chrX_+_70232362
|
0.888
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr3_-_15081705
|
0.887
|
|
MRPS25
|
mitochondrial ribosomal protein S25
|
|
chr19_+_62638474
|
0.885
|
NM_001023561
|
ZNF749
|
zinc finger protein 749
|
|
chr11_+_133628616
|
0.879
|
NM_014384
|
ACAD8
|
acyl-CoA dehydrogenase family, member 8
|
|
chr4_-_2934834
|
0.872
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr17_+_35550032
|
0.870
|
NM_007359
|
CASC3
|
cancer susceptibility candidate 3
|
|
chr2_-_178125680
|
0.869
|
NM_152517
|
TTC30B
|
tetratricopeptide repeat domain 30B
|
|
chr14_-_22468393
|
0.869
|
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr19_-_19748186
|
0.866
|
|
LOC284440
|
hypothetical LOC284440
|
|
chr15_-_50648469
|
0.857
|
NM_006628
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr17_+_7532391
|
0.856
|
NM_001143992
NM_018081
|
WRAP53
|
WD repeat containing, antisense to TP53
|
|
chr12_+_54784600
|
0.855
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr14_-_22468445
|
0.847
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr19_+_14412052
|
0.847
|
|
PKN1
|
protein kinase N1
|
|
chr3_+_23962518
|
0.845
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr14_+_64523700
|
0.845
|
|
|
|
|
chr17_+_5264195
|
0.842
|
|
RPAIN
|
RPA interacting protein
|
|
chr11_+_77577505
|
0.836
|
NM_020798
|
USP35
|
ubiquitin specific peptidase 35
|
|
chr19_+_49290319
|
0.835
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr11_-_65244718
|
0.831
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr19_+_55844446
|
0.826
|
NM_001195076
|
LOC342918
|
hypothetical protein LOC342918
|
|
chr4_-_5879488
|
0.825
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr16_+_4415805
|
0.824
|
NM_001135110
NM_005147
|
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3
|
|
chr3_-_43122474
|
0.823
|
NM_032806
|
C3orf39
|
chromosome 3 open reading frame 39
|
|
chr10_-_42368035
|
0.821
|
|
ZNF37BP
|
zinc finger protein 37B, pseudogene
|
|
chr12_-_49763405
|
0.820
|
|
CSRNP2
|
cysteine-serine-rich nuclear protein 2
|
|
chr12_+_54784591
|
0.820
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr2_+_172252473
|
0.802
|
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr19_+_14412080
|
0.793
|
NM_213560
|
PKN1
|
protein kinase N1
|
|
chr11_-_60885849
|
0.779
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr15_+_38886546
|
0.770
|
NM_001077268
|
ZFYVE19
|
zinc finger, FYVE domain containing 19
|
|
chr3_+_170973870
|
0.769
|
|
MYNN
|
myoneurin
|
|
chr19_+_44082098
|
0.768
|
|
|
|
|
chr6_+_109523388
|
0.768
|
|
CEP57L1
|
centrosomal protein 57kDa-like 1
|
|
chr9_-_19092648
|
0.767
|
|
HAUS6
|
HAUS augmin-like complex, subunit 6
|
|
chr20_-_1395463
|
0.765
|
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chr22_-_28492990
|
0.761
|
|
|
|
|
chr9_-_101900945
|
0.760
|
|
ERP44
|
endoplasmic reticulum protein 44
|
|
chr21_-_46703020
|
0.760
|
|
|
|
|
chr12_-_94953427
|
0.758
|
NM_000895
|
LTA4H
|
leukotriene A4 hydrolase
|
|
chr11_-_73559507
|
0.758
|
NM_015531
|
C2CD3
|
C2 calcium-dependent domain containing 3
|
|
chr19_+_59297953
|
0.758
|
NM_004542
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
|
|
chr19_-_47438518
|
0.753
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr20_-_34807882
|
0.752
|
NM_022477
NM_032013
|
NDRG3
|
NDRG family member 3
|
|
chr22_-_40012133
|
0.751
|
NM_002883
|
RANGAP1
|
Ran GTPase activating protein 1
|
|
chr19_+_14412087
|
0.749
|
|
PKN1
|
protein kinase N1
|
|
chr21_-_26029830
|
0.746
|
NM_001003696
NM_001003697
NM_001003701
NM_001003703
NM_001685
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6
|
|
chr20_-_48165886
|
0.745
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr7_+_150356435
|
0.741
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr5_-_171366481
|
0.738
|
NM_012300
NM_033644
NM_033645
|
FBXW11
|
F-box and WD repeat domain containing 11
|
|
chr12_+_54784614
|
0.736
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr21_-_37562060
|
0.733
|
|
DSCR3
|
Down syndrome critical region gene 3
|
|
chr7_+_89870647
|
0.729
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr19_-_56303434
|
0.724
|
NM_145232
|
CTU1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe)
|
|
chr13_-_32010883
|
0.723
|
NM_014887
NM_033111
|
N4BP2L2
|
NEDD4 binding protein 2-like 2
|
|
chr9_-_103200697
|
0.722
|
NM_019051
|
MRPL50
|
mitochondrial ribosomal protein L50
|
|
chr6_+_2710675
|
0.719
|
|
WRNIP1
|
Werner helicase interacting protein 1
|
|
chr16_-_2767129
|
0.718
|
NM_007108
NM_207013
|
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B)
|
|
chr14_-_76856776
|
0.717
|
NM_013382
|
POMT2
|
protein-O-mannosyltransferase 2
|
|
chr12_-_111304138
|
0.716
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr12_+_54784625
|
0.715
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr7_-_74994844
|
0.714
|
|
PMS2P3
|
postmeiotic segregation increased 2 pseudogene 3
|
|
chr3_+_4320036
|
0.713
|
|
SETMAR
|
SET domain and mariner transposase fusion gene
|
|
chr1_-_10455100
|
0.711
|
|
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide
|
|
chr2_-_178645718
|
0.710
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr1_-_21478571
|
0.709
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_+_218896102
|
0.708
|
NM_022572
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr12_-_119938593
|
0.706
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr17_+_28279040
|
0.706
|
NM_001033504
NM_015544
|
TMEM98
|
transmembrane protein 98
|
|
chr11_+_82460766
|
0.705
|
|
LOC100506233
|
hypothetical LOC100506233
|
|
chr2_+_24869753
|
0.698
|
NM_024322
|
CENPO
|
centromere protein O
|
|
chr16_-_3225360
|
0.697
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr4_-_2934945
|
0.696
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr13_+_49554307
|
0.695
|
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding)
|
|
chr7_-_134845363
|
0.693
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr19_-_44082008
|
0.691
|
NM_012237
NM_001193286
NM_030593
|
SIRT2
|
sirtuin 2
|
|
chr6_+_30983941
|
0.688
|
NM_001517
|
GTF2H4
|
general transcription factor IIH, polypeptide 4, 52kDa
|
|
chr16_-_31013423
|
0.688
|
|
|
|
|
chr18_+_69966725
|
0.684
|
NM_014177
|
C18orf55
|
chromosome 18 open reading frame 55
|
|
chr15_+_38240501
|
0.684
|
NM_001211
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr4_-_47611255
|
0.683
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr6_+_36518778
|
0.683
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr4_-_2934909
|
0.681
|
NM_003703
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr16_-_3124859
|
0.677
|
|
|
|
|
chr2_+_232354675
|
0.677
|
|
COPS7B
|
COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis)
|
|
chr2_+_218843358
|
0.675
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr11_-_75833337
|
0.673
|
|
|
|
|
chr6_+_42639929
|
0.671
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr1_-_28780940
|
0.666
|
|
SNHG12
|
small nucleolar RNA host gene 12 (non-protein coding)
|
|
chr11_+_16716751
|
0.666
|
|
C11orf58
|
chromosome 11 open reading frame 58
|
|
chr7_+_128651976
|
0.661
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr4_-_2934889
|
0.659
|
|
NOP14
|
NOP14 nucleolar protein homolog (yeast)
|
|
chr20_+_30259359
|
0.659
|
|
POFUT1
|
protein O-fucosyltransferase 1
|
|
chr11_-_82460531
|
0.656
|
NM_014488
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr21_-_34209897
|
0.656
|
NM_001697
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit
|