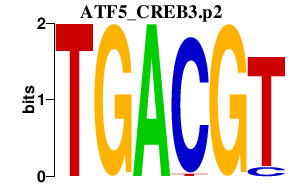
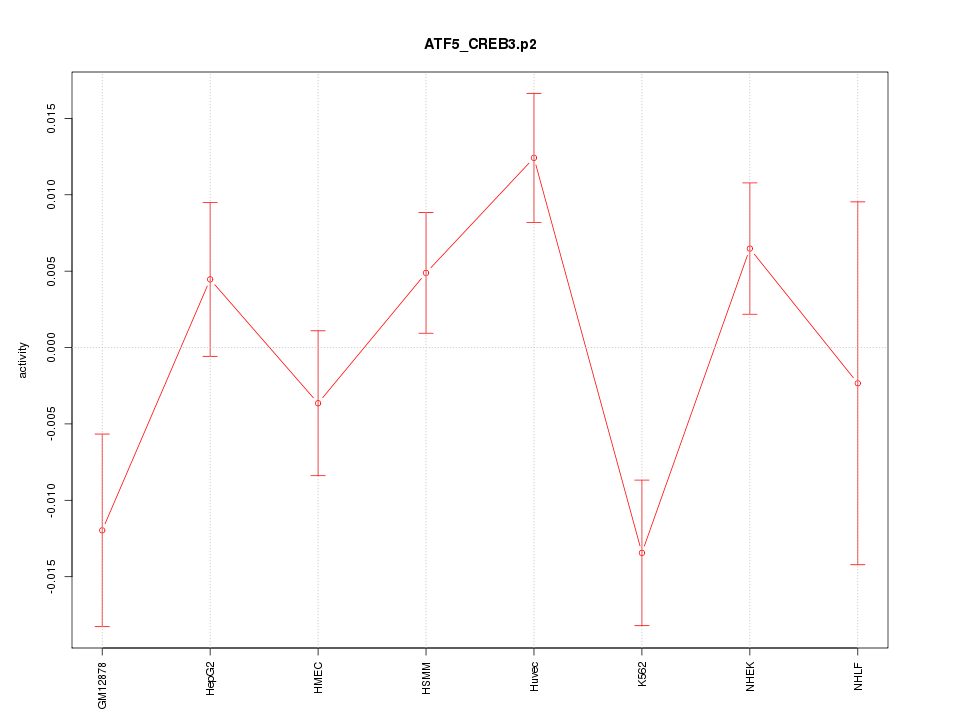
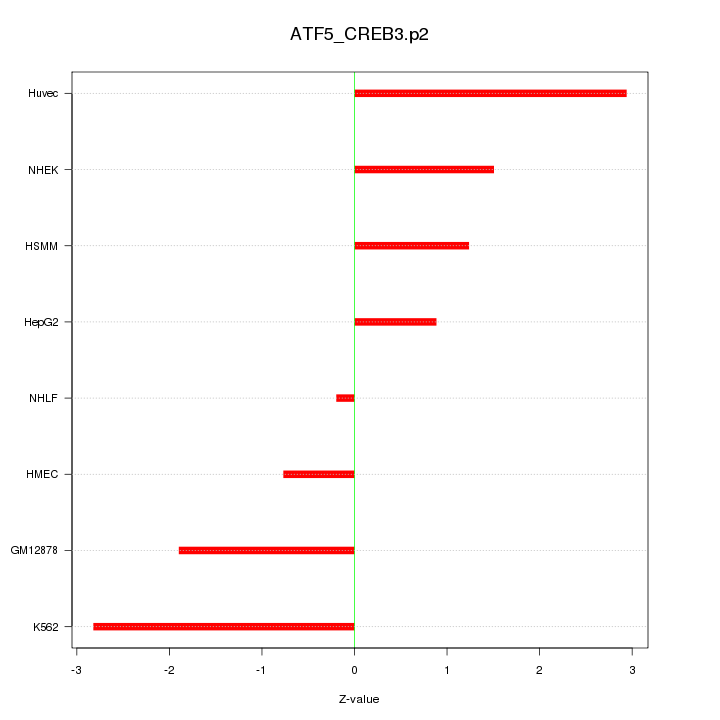
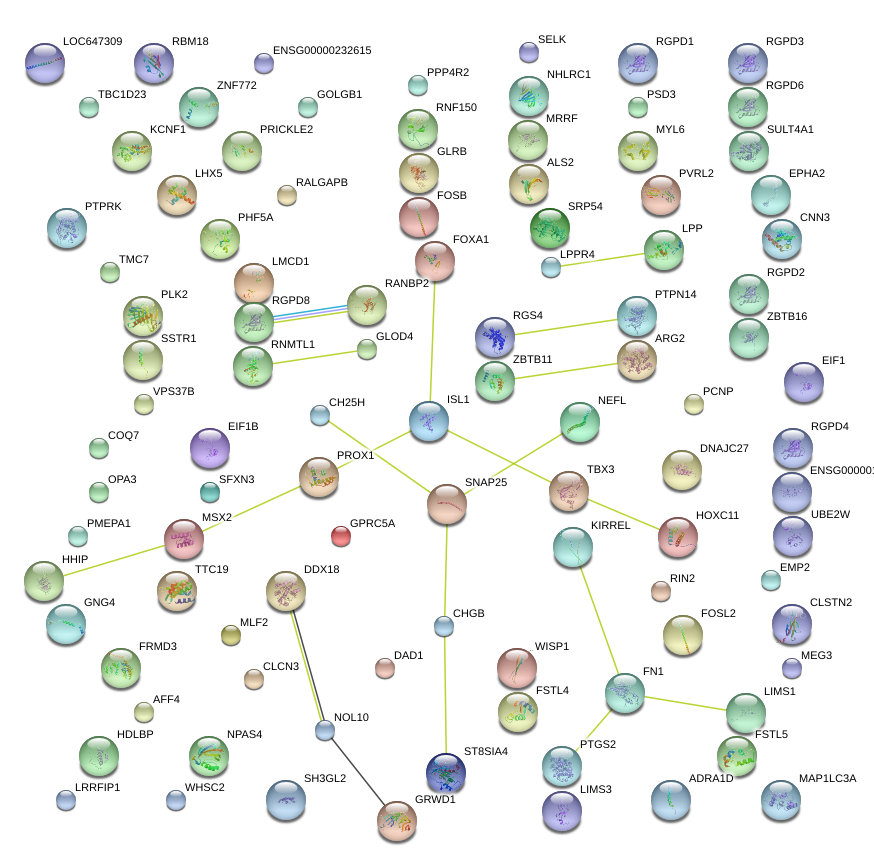
|
chr20_+_32610161
|
3.114
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr3_-_102878301
|
2.846
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr4_-_187884721
|
2.010
|
|
|
|
|
chr3_+_141136691
|
1.522
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr2_+_10969513
|
1.436
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr22_-_42589543
|
1.422
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr3_+_102775677
|
1.361
|
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr2_+_108702338
|
1.302
|
NM_006267
|
RANBP2
|
RAN binding protein 2
|
|
chr3_+_101462375
|
1.284
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr4_+_145786622
|
1.172
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr14_+_67156323
|
1.100
|
NM_001172
|
ARG2
|
arginase, type II
|
|
chr17_+_37098668
|
1.050
|
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr3_+_141136896
|
1.001
|
|
CLSTN2
|
calsyntenin 2
|
|
chr2_-_216009015
|
0.993
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr1_-_212791597
|
0.969
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr20_-_4177519
|
0.968
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr17_+_632356
|
0.948
|
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr4_-_1980756
|
0.939
|
NM_005663
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr3_-_64186151
|
0.939
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_-_212791467
|
0.937
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr10_+_102782151
|
0.900
|
|
SFXN3
|
sideroflexin 3
|
|
chr6_-_128883125
|
0.885
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr12_+_54838408
|
0.876
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr16_+_18986721
|
0.852
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr2_+_238200922
|
0.850
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr1_+_99502655
|
0.848
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr12_+_52653176
|
0.847
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr19_-_50779861
|
0.844
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr12_-_6732392
|
0.838
|
|
MLF2
|
myeloid leukemia factor 2
|
|
chr5_+_50714996
|
0.836
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr1_-_233879617
|
0.820
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr9_+_17568952
|
0.814
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr19_-_50779914
|
0.809
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr5_-_57791544
|
0.808
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr5_-_50714836
|
0.800
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr2_+_118288754
|
0.796
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr2_+_118288724
|
0.791
|
NM_006773
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr10_-_90957028
|
0.789
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr2_+_118288753
|
0.786
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr3_-_122951200
|
0.766
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr11_+_65945050
|
0.756
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr3_-_189354510
|
0.751
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr1_-_20682541
|
0.737
|
|
|
|
|
chr3_-_53900888
|
0.737
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr3_-_8518327
|
0.734
|
|
LOC100288428
|
hypothetical LOC100288428
|
|
chr6_-_18230829
|
0.733
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr3_+_8518508
|
0.732
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr14_+_37748031
|
0.731
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr3_+_8518539
|
0.730
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr12_+_54838361
|
0.727
|
NM_021019
NM_079423
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr16_+_18986417
|
0.727
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr20_+_19903778
|
0.727
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr19_+_50041400
|
0.724
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr2_-_202353898
|
0.723
|
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr9_-_85342868
|
0.722
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr2_+_28469172
|
0.719
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr5_-_132326872
|
0.702
|
|
AFF4
|
AF4/FMR2 family, member 4
|
|
chr20_+_19863716
|
0.699
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr14_+_34521854
|
0.695
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr22_-_40194602
|
0.691
|
NM_032758
|
PHF5A
|
PHD finger protein 5A
|
|
chr3_+_102775716
|
0.688
|
NM_020357
|
PCNP
|
PEST proteolytic signal containing nuclear protein
|
|
chr20_+_10147455
|
0.685
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr1_-_95165037
|
0.684
|
|
CNN3
|
calponin 3, acidic
|
|
chr19_+_50041387
|
0.682
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr3_+_189354167
|
0.671
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr14_-_22127954
|
0.665
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr14_-_22127902
|
0.663
|
|
DAD1
|
defender against cell death 1
|
|
chr1_+_212227902
|
0.659
|
|
PROX1
|
prospero homeobox 1
|
|
chr12_-_121946581
|
0.659
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr20_+_5839973
|
0.657
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr1_-_95165089
|
0.656
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr4_-_163304567
|
0.651
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chr12_+_12935619
|
0.650
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr2_-_25048276
|
0.646
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr8_-_18915443
|
0.644
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr14_+_100362236
|
0.631
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr12_-_113606322
|
0.629
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr9_-_124066901
|
0.626
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr9_+_124066973
|
0.618
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr19_-_50779924
|
0.616
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr17_+_632262
|
0.616
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr20_+_36534871
|
0.616
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr2_-_216008689
|
0.612
|
|
FN1
|
fibronectin 1
|
|
chr5_-_132976094
|
0.609
|
|
FSTL4
|
follistatin-like 4
|
|
chr8_-_74953626
|
0.606
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr19_+_50663089
|
0.601
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr17_-_632135
|
0.601
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr19_+_53641053
|
0.601
|
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr8_-_74953604
|
0.600
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr2_+_108637916
|
0.599
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr11_+_113435640
|
0.596
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr19_-_62680703
|
0.595
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr4_-_142273881
|
0.594
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr17_+_15843418
|
0.593
|
NM_017775
|
TTC19
|
tetratricopeptide repeat domain 19
|
|
chr8_-_74953632
|
0.592
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr1_-_16355013
|
0.591
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr2_+_108637698
|
0.590
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_+_50714714
|
0.590
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr19_+_53641029
|
0.589
|
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr5_-_100266769
|
0.585
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr12_+_12935222
|
0.582
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr19_+_50041424
|
0.579
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr8_-_24869945
|
0.579
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr4_+_170778265
|
0.575
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr12_-_6732342
|
0.574
|
NM_005439
|
MLF2
|
myeloid leukemia factor 2
|
|
chr14_+_100362204
|
0.567
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr16_+_18986459
|
0.565
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_-_192063158
|
0.564
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr5_-_132976121
|
0.564
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr20_-_55718351
|
0.562
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr2_-_241860871
|
0.557
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr12_+_12935759
|
0.554
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr14_-_37134191
|
0.552
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr5_+_174084137
|
0.550
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr12_-_112394259
|
0.549
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr2_-_27967458
|
0.549
|
|
LOC100302650
|
hypothetical LOC100302650
|
|
chr1_+_156229934
|
0.548
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr2_-_202354139
|
0.547
|
NM_001135745
NM_020919
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr8_+_134272463
|
0.547
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr1_+_161305558
|
0.547
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr8_-_74953648
|
0.546
|
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr16_+_18902756
|
0.544
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr16_-_10581944
|
0.540
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr3_+_73128771
|
0.539
|
NM_174907
|
PPP4R2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr11_+_113435506
|
0.537
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr1_-_184916044
|
0.533
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr2_-_10747553
|
0.533
|
NM_024894
|
NOL10
|
nucleolar protein 10
|
|
chr4_+_158216631
|
0.532
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr2_-_10747438
|
0.530
|
|
NOL10
|
nucleolar protein 10
|
|
chr6_+_30702586
|
0.529
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr2_+_28469275
|
0.526
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr11_+_12652427
|
0.525
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr2_+_28469199
|
0.523
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr9_-_74169649
|
0.522
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr9_-_124066838
|
0.513
|
|
RBM18
|
RNA binding motif protein 18
|
|
chrX_+_43399101
|
0.512
|
|
MAOA
|
monoamine oxidase A
|
|
chr2_-_241860923
|
0.510
|
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr14_+_34521945
|
0.509
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr12_-_55013987
|
0.503
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr9_+_115023628
|
0.502
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr6_+_148705817
|
0.502
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr15_+_91248706
|
0.499
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr12_-_45052830
|
0.497
|
NM_018976
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr5_-_37875538
|
0.496
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr8_-_74953652
|
0.495
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr8_-_11096257
|
0.490
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr9_-_32991518
|
0.490
|
NM_001195249
NM_001195254
NM_001195248
NM_001195250
NM_001195251
NM_001195252
NM_175069
NM_175073
|
APTX
|
aprataxin
|
|
chr22_+_36927884
|
0.487
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr9_-_26882713
|
0.485
|
NM_001167575
NM_024828
|
C9orf82
|
chromosome 9 open reading frame 82
|
|
chr7_+_71937980
|
0.484
|
|
SBDSP1
|
Shwachman-Bodian-Diamond syndrome pseudogene 1
|
|
chr4_-_8481085
|
0.484
|
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr19_+_53814602
|
0.482
|
|
SPHK2
|
sphingosine kinase 2
|
|
chr1_-_208046011
|
0.481
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr2_+_5750229
|
0.478
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr10_+_102782156
|
0.477
|
|
SFXN3
|
sideroflexin 3
|
|
chr4_-_111777584
|
0.477
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr13_-_20533629
|
0.477
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr9_+_91123204
|
0.474
|
NM_024077
|
SECISBP2
|
SECIS binding protein 2
|
|
chr4_-_78216145
|
0.473
|
NM_006835
|
CCNI
|
cyclin I
|
|
chr13_-_20533703
|
0.471
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr6_+_30693464
|
0.471
|
NM_014046
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr14_+_23905984
|
0.469
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr2_+_110013297
|
0.468
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr17_-_20739924
|
0.467
|
NM_001004306
|
CCDC144NL
|
coiled-coil domain containing 144 family, N-terminal like
|
|
chr12_-_123568781
|
0.467
|
|
|
|
|
chr4_+_146238936
|
0.467
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr13_-_27967215
|
0.466
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr2_-_202353818
|
0.466
|
|
|
|
|
chr2_-_191723951
|
0.465
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr5_+_121675672
|
0.460
|
NM_005460
|
SNCAIP
|
synuclein, alpha interacting protein
|
|
chr16_+_18903009
|
0.460
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr14_+_99017485
|
0.459
|
NM_001099402
|
CCNK
|
cyclin K
|
|
chr14_-_49122813
|
0.458
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr6_+_39868116
|
0.458
|
|
|
|
|
chr12_-_47604930
|
0.458
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr4_+_146238818
|
0.457
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr13_-_32010883
|
0.452
|
NM_014887
NM_033111
|
N4BP2L2
|
NEDD4 binding protein 2-like 2
|
|
chr2_+_131579355
|
0.452
|
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2
|
|
chr2_+_190916571
|
0.451
|
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr17_+_8093319
|
0.449
|
NM_012393
|
PFAS
|
phosphoribosylformylglycinamidine synthase
|
|
chr9_+_111582317
|
0.449
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr3_+_134775094
|
0.448
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr19_+_53814574
|
0.447
|
|
SPHK2
|
sphingosine kinase 2
|
|
chr17_+_37098652
|
0.447
|
NM_005801
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr14_-_38971370
|
0.447
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr14_+_67156386
|
0.445
|
|
ARG2
|
arginase, type II
|
|
chr12_-_55013713
|
0.445
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr4_-_146238746
|
0.444
|
NM_014885
|
ANAPC10
|
anaphase promoting complex subunit 10
|
|
chr12_-_10657397
|
0.442
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr4_-_44145580
|
0.442
|
NM_198353
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr17_-_46553083
|
0.441
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr11_-_103540275
|
0.440
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr4_-_111777651
|
0.440
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr17_+_37098667
|
0.439
|
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr5_-_148911105
|
0.438
|
|
CSNK1A1
|
casein kinase 1, alpha 1
|
|
chr21_+_32706615
|
0.437
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr3_-_151746589
|
0.436
|
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr3_-_151963607
|
0.434
|
|
SIAH2
|
seven in absentia homolog 2 (Drosophila)
|
|
chr15_+_60146467
|
0.432
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr5_-_59100194
|
0.431
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|