|
chr5_-_160044844
|
1.048
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_-_11313907
|
0.846
|
NM_006249
|
PRB3
|
proline-rich protein BstNI subfamily 3
|
|
chr1_+_50344550
|
0.695
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_+_22646229
|
0.602
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr5_+_127012611
|
0.591
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr4_-_72868598
|
0.529
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr1_+_57093030
|
0.494
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr14_+_87560646
|
0.425
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr11_+_59038961
|
0.424
|
NM_001004711
|
OR4D9
|
olfactory receptor, family 4, subfamily D, member 9
|
|
chr3_-_15773061
|
0.417
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr9_+_109131814
|
0.417
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr1_-_156078115
|
0.416
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr14_-_63076155
|
0.403
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr19_-_63556632
|
0.398
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr12_+_109956210
|
0.391
|
NM_015267
|
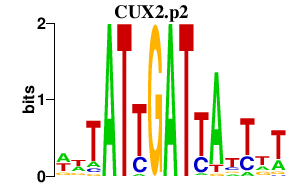
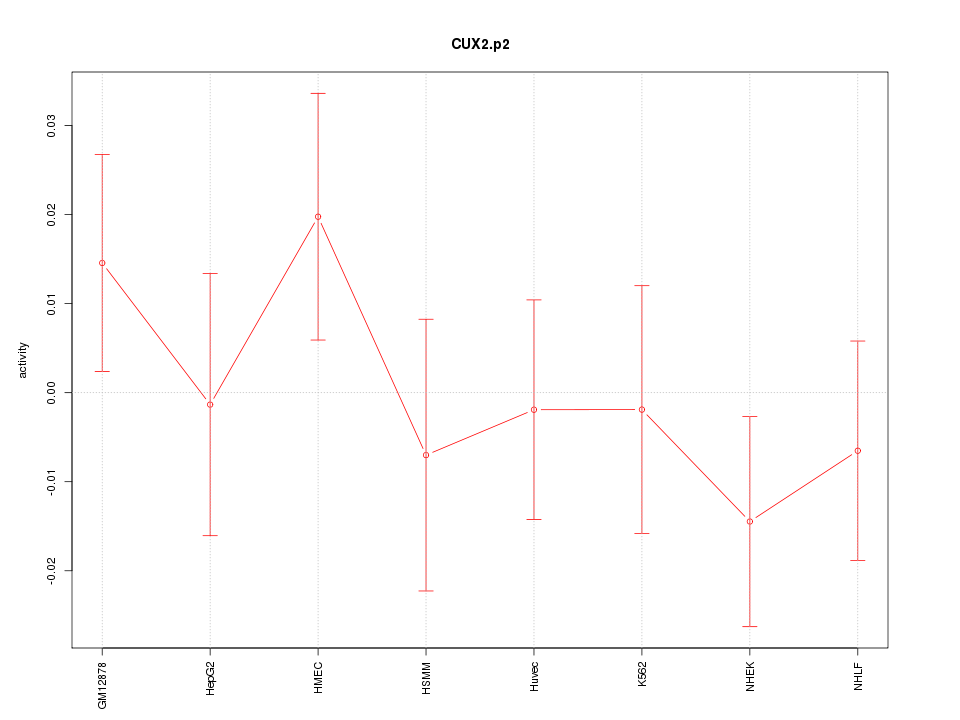
CUX2
|
cut-like homeobox 2
|
|
chr7_-_36730525
|
0.377
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr9_-_106420305
|
0.373
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr3_+_85858321
|
0.367
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr3_+_161040439
|
0.365
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr6_-_161614962
|
0.348
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr11_-_77411933
|
0.321
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr3_+_153499883
|
0.311
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr9_+_5646582
|
0.309
|
NM_020829
|
KIAA1432
|
KIAA1432
|
|
chr12_-_53661888
|
0.306
|
NM_001098815
|
KIAA0748
|
KIAA0748
|
|
chr14_-_23090606
|
0.301
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr7_+_80069439
|
0.289
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_+_161040343
|
0.288
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr22_-_25291268
|
0.286
|
NM_001008566
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr20_-_33705244
|
0.263
|
NM_003915
|
CPNE1
|
copine I
|
|
chr8_+_18293034
|
0.261
|
NM_000015
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr1_-_144779601
|
0.260
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr13_-_35327798
|
0.255
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr22_+_23921072
|
0.240
|
|
KIAA1671
|
KIAA1671
|
|
chr12_+_54011750
|
0.219
|
NM_054104
|
OR6C3
|
olfactory receptor, family 6, subfamily C, member 3
|
|
chr5_+_36642213
|
0.201
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_-_52939636
|
0.195
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr2_-_178461710
|
0.191
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr1_+_246635918
|
0.179
|
NM_030904
|
OR2T1
|
olfactory receptor, family 2, subfamily T, member 1
|
|
chr4_-_70861301
|
0.178
|
NM_001891
|
CSN2
|
casein beta
|
|
chr3_+_63403792
|
0.171
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr1_-_146076704
|
0.171
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr2_+_108230082
|
0.171
|
NM_001008743
|
SULT1C3
|
sulfotransferase family, cytosolic, 1C, member 3
|
|
chr16_+_11966464
|
0.168
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr4_+_77214886
|
0.153
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr1_+_110795271
|
0.149
|
NM_032414
|
PROK1
|
prokineticin 1
|
|
chr4_+_71126376
|
0.147
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr6_+_46869052
|
0.144
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr7_+_156677777
|
0.144
|
|
|
|
|
chr17_-_61655915
|
0.137
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr7_+_143646150
|
0.134
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chrX_-_117003310
|
0.128
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr9_-_5294579
|
0.127
|
NM_005059
NM_134441
|
RLN2
|
relaxin 2
|
|
chr12_-_52939584
|
0.124
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr4_+_71330797
|
0.118
|
NM_001145006
|
MUC7
|
mucin 7, secreted
|
|
chr14_+_21203136
|
0.115
|
NM_001001912
|
OR4E2
|
olfactory receptor, family 4, subfamily E, member 2
|
|
chr9_+_27099138
|
0.115
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_-_27907283
|
0.112
|
NM_003541
|
HIST1H4A
HIST1H4F
HIST1H4K
HIST1H4J
|
histone cluster 1, H4a
histone cluster 1, H4f
histone cluster 1, H4k
histone cluster 1, H4j
|
|
chr10_-_98326435
|
0.111
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr12_+_6703410
|
0.108
|
NM_001164094
NM_016319
NM_001164095
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr4_-_68766392
|
0.106
|
NM_001129907
|
TMPRSS11BNL
|
TMPRSS11B N terminal-like
|
|
chr10_+_90474280
|
0.106
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr7_-_143560850
|
0.106
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr5_+_177415181
|
0.105
|
|
|
|
|
chr2_-_98646367
|
0.103
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr14_-_73095396
|
0.100
|
NM_203309
|
HEATR4
|
HEAT repeat containing 4
|
|
chr21_+_30690165
|
0.099
|
NM_181599
|
KRTAP13-1
|
keratin associated protein 13-1
|
|
chr12_+_6424293
|
0.096
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr17_+_53670785
|
0.095
|
NM_001160102
NM_006151
|
LPO
|
lactoperoxidase
|
|
chr4_-_47678187
|
0.095
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr14_-_20098929
|
0.095
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr17_+_65009986
|
0.095
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr2_+_169634330
|
0.094
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr11_+_60016826
|
0.092
|
NM_001164470
NM_017716
|
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12
|
|
chr12_-_10870134
|
0.089
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr11_+_33306652
|
0.088
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr1_+_144374513
|
0.087
|
|
RNF115
|
ring finger protein 115
|
|
chr12_+_21059896
|
0.086
|
NM_001009562
|
LST-3TM12
|
organic anion transporter LST-3b
|
|
chrX_-_11194015
|
0.086
|
NM_013423
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr7_+_99263571
|
0.078
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr12_+_6703549
|
0.078
|
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr12_-_27815475
|
0.073
|
NM_001146221
|
LOC100287284
|
MANSC domain-containing protein ENSP00000370673
|
|
chr19_-_53935648
|
0.073
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chr18_+_59767567
|
0.071
|
NM_001123366
|
HMSD
|
histocompatibility (minor) serpin domain containing
|
|
chr3_-_150422474
|
0.070
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr8_-_120720200
|
0.070
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr5_-_58918031
|
0.070
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_23476497
|
0.070
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr5_-_43448174
|
0.069
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr7_-_91632525
|
0.068
|
NM_001161528
|
LOC401387
|
leucine-rich repeat and death domain-containing protein
|
|
chr5_-_94250357
|
0.067
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr11_-_34489693
|
0.067
|
NM_198381
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr12_-_21980894
|
0.066
|
NM_005691
NM_020297
NM_020298
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr5_+_57823017
|
0.065
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr11_-_57964221
|
0.065
|
NM_001004733
|
OR5B12
|
olfactory receptor, family 5, subfamily B, member 12
|
|
chr7_+_116238309
|
0.063
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr11_-_74478405
|
0.062
|
NM_001005285
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4
|
|
chr17_-_8364942
|
0.062
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr7_-_18033970
|
0.061
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr18_-_29882555
|
0.060
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr14_-_23117268
|
0.058
|
|
JPH4
|
junctophilin 4
|
|
chr13_-_45654296
|
0.057
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chrX_+_30158473
|
0.055
|
NM_002365
|
MAGEB3
|
melanoma antigen family B, 3
|
|
chr5_+_140509803
|
0.055
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr8_-_82557956
|
0.055
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr3_-_172227152
|
0.053
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr5_+_140227951
|
0.053
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr3_-_127760447
|
0.052
|
NM_152533
|
C3orf22
|
chromosome 3 open reading frame 22
|
|
chr11_+_59902530
|
0.052
|
NM_021201
NM_206938
NM_206939
NM_206940
|
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7
|
|
chrX_-_118610880
|
0.052
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr14_-_19867372
|
0.052
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr6_-_68655708
|
0.052
|
|
|
|
|
chr8_+_11225904
|
0.052
|
NM_054028
|
AMAC1L2
|
acyl-malonyl condensing enzyme 1-like 2
|
|
chr2_+_114935623
|
0.052
|
NM_001178036
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr12_-_116890410
|
0.051
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr15_-_70838703
|
0.049
|
|
|
|
|
chr17_-_2943039
|
0.049
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr1_-_245942679
|
0.049
|
NM_001005286
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1
|
|
chr11_+_5732498
|
0.047
|
NM_001005175
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4
|
|
chr3_-_42427026
|
0.047
|
NM_144634
|
LYZL4
|
lysozyme-like 4
|
|
chr12_-_11354620
|
0.047
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr1_+_357502
|
0.047
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr12_+_73217817
|
0.045
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr21_-_30460841
|
0.045
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr11_+_55899675
|
0.043
|
NM_001005204
NM_001013356
|
OR8U1
OR8U8
|
olfactory receptor, family 8, subfamily U, member 1
olfactory receptor, family 8, subfamily U, member 8
|
|
chr3_-_187744808
|
0.039
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chrX_-_15312390
|
0.038
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr2_-_162717159
|
0.038
|
NM_002054
|
GCG
|
glucagon
|
|
chr10_-_98280299
|
0.036
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr10_+_104580420
|
0.036
|
|
|
|
|
chrX_+_106057984
|
0.035
|
|
CLDN2
|
claudin 2
|
|
chr11_+_55407348
|
0.035
|
NM_032681
|
SPRYD5
|
SPRY domain containing 5
|
|
chr18_+_57308754
|
0.034
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr1_+_51340487
|
0.034
|
NM_001136508
|
C1orf185
|
chromosome 1 open reading frame 185
|
|
chr5_+_121493113
|
0.033
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr15_-_64582049
|
0.032
|
|
RPL4
|
ribosomal protein L4
|
|
chr11_+_59902598
|
0.032
|
|
MS4A7
|
membrane-spanning 4-domains, subfamily A, member 7
|
|
chr12_+_102859354
|
0.032
|
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr1_+_204797079
|
0.030
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr15_-_77169894
|
0.030
|
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr6_+_31081707
|
0.029
|
NM_001198815
|
PBMUCL1
|
panbronchiolitis related mucin-like 1
|
|
chr2_-_171999473
|
0.029
|
NM_024770
|
METTL8
|
methyltransferase like 8
|
|
chr2_-_20375497
|
0.028
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr3_+_44667504
|
0.028
|
|
ZNF35
|
zinc finger protein 35
|
|
chr11_+_91724909
|
0.027
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr10_+_69539255
|
0.027
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr6_-_26216299
|
0.027
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chrX_+_30169977
|
0.026
|
NM_002367
|
MAGEB4
|
melanoma antigen family B, 4
|
|
chr11_+_61794205
|
0.025
|
NM_002411
|
SCGB2A2
|
secretoglobin, family 2A, member 2
|
|
chr12_+_6703525
|
0.025
|
|
COPS7A
|
COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis)
|
|
chr3_+_173043832
|
0.025
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr6_+_32817140
|
0.025
|
NM_020056
|
HLA-DQA2
|
major histocompatibility complex, class II, DQ alpha 2
|
|
chr18_-_62422195
|
0.025
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr12_+_26017954
|
0.024
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr20_-_22514092
|
0.023
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr3_-_172227461
|
0.022
|
NM_000340
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr12_+_124377114
|
0.021
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr13_-_45654327
|
0.021
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr3_+_120983556
|
0.021
|
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr4_-_47160476
|
0.021
|
|
COMMD8
|
COMM domain containing 8
|
|
chr17_+_7402315
|
0.021
|
NM_001198622
NM_001198623
NM_001198624
NM_003808
NM_172087
NM_172088
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chrX_+_134482212
|
0.020
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr18_-_68683913
|
0.020
|
NM_138999
NM_153181
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr11_+_60223622
|
0.019
|
NM_031457
|
MS4A8B
|
membrane-spanning 4-domains, subfamily A, member 8B
|
|
chr3_+_159310631
|
0.019
|
|
RSRC1
|
arginine/serine-rich coiled-coil 1
|
|
chr3_-_172227223
|
0.019
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr3_+_51837608
|
0.018
|
NM_001085479
|
IQCF3
|
IQ motif containing F3
|
|
chr9_-_21177529
|
0.018
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr1_+_165330710
|
0.018
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr7_+_93373755
|
0.018
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr10_-_93992982
|
0.016
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr12_-_15757114
|
0.015
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr2_+_69055208
|
0.015
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr2_-_152538694
|
0.014
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr13_+_50813168
|
0.014
|
NM_001101320
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3
|
|
chr17_-_44042924
|
0.013
|
|
HOXB7
|
homeobox B7
|
|
chr8_+_67505091
|
0.013
|
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae)
|
|
chr11_+_57121602
|
0.013
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr7_+_116182676
|
0.013
|
|
|
|
|
chr1_-_150805852
|
0.013
|
NM_178435
|
LCE3E
|
late cornified envelope 3E
|
|
chr3_+_135996734
|
0.012
|
NM_004441
|
EPHB1
|
EPH receptor B1
|
|
chr12_-_7709734
|
0.012
|
NM_001644
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
|
|
chr11_-_30562324
|
0.011
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr1_-_611896
|
0.011
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr10_-_69095362
|
0.011
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr22_-_38619739
|
0.011
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chrX_+_10084984
|
0.010
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr5_+_60969392
|
0.010
|
NM_173667
|
FLJ37543
|
hypothetical protein FLJ37543
|
|
chr6_+_116956868
|
0.009
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr4_+_41395493
|
0.009
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr15_+_36014748
|
0.009
|
NM_152453
|
TMCO5A
|
transmembrane and coiled-coil domains 5A
|
|
chr11_-_5766621
|
0.008
|
NM_001001913
|
OR52N1
|
olfactory receptor, family 52, subfamily N, member 1
|
|
chr1_+_117098579
|
0.008
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr1_-_120285753
|
0.007
|
|
NOTCH2
|
notch 2
|
|
chr4_-_82345224
|
0.007
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|
|
chr21_+_29322100
|
0.007
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr9_-_129537372
|
0.006
|
NM_001085347
NM_001134430
NM_001134431
NM_130459
|
TOR2A
|
torsin family 2, member A
|
|
chr4_+_187424111
|
0.006
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr7_-_112515068
|
0.005
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chrX_+_123307812
|
0.005
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr10_-_69125932
|
0.004
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|