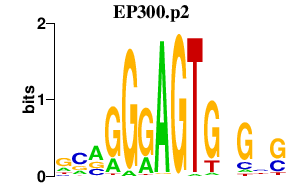
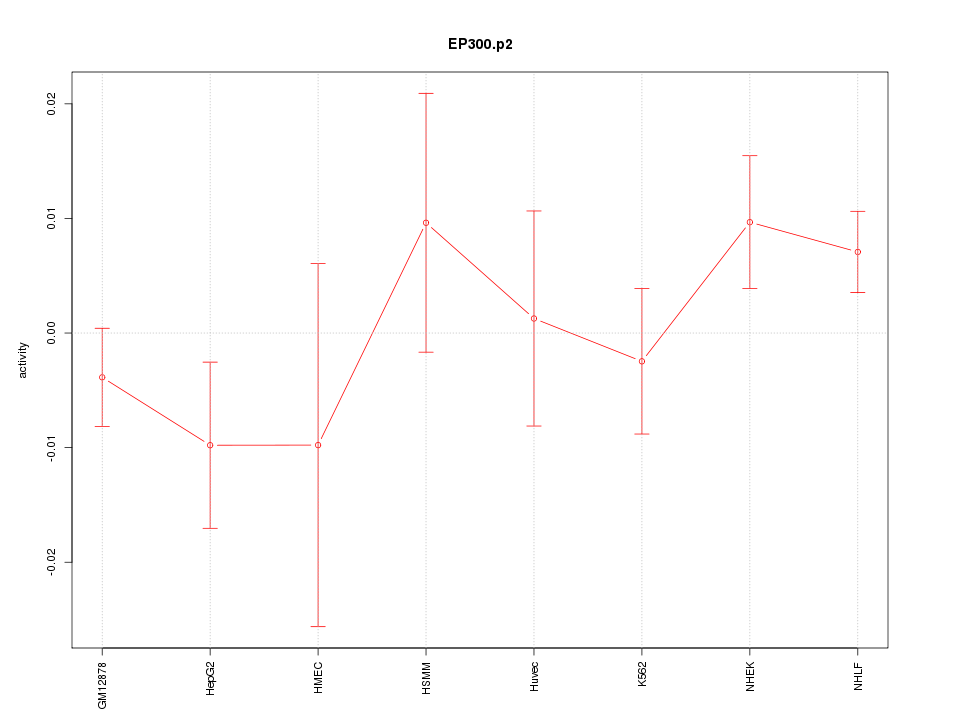
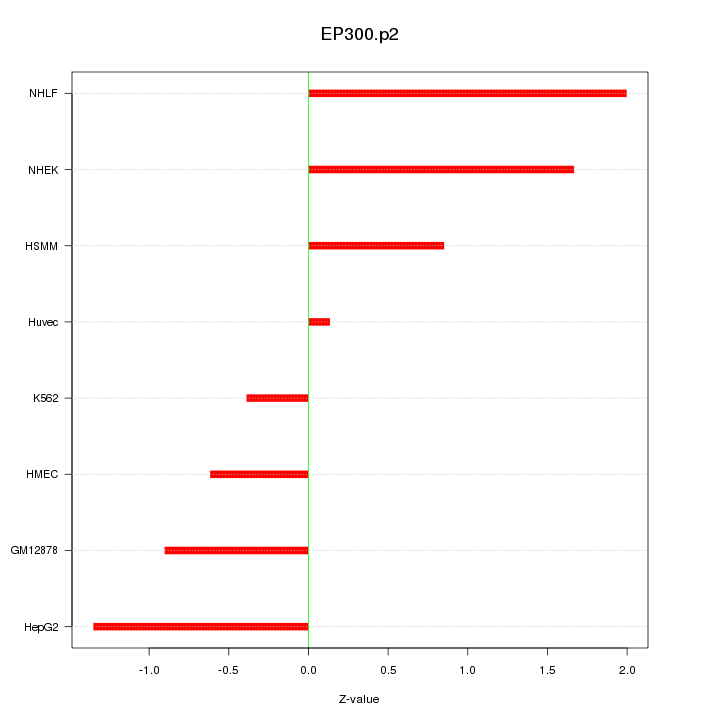
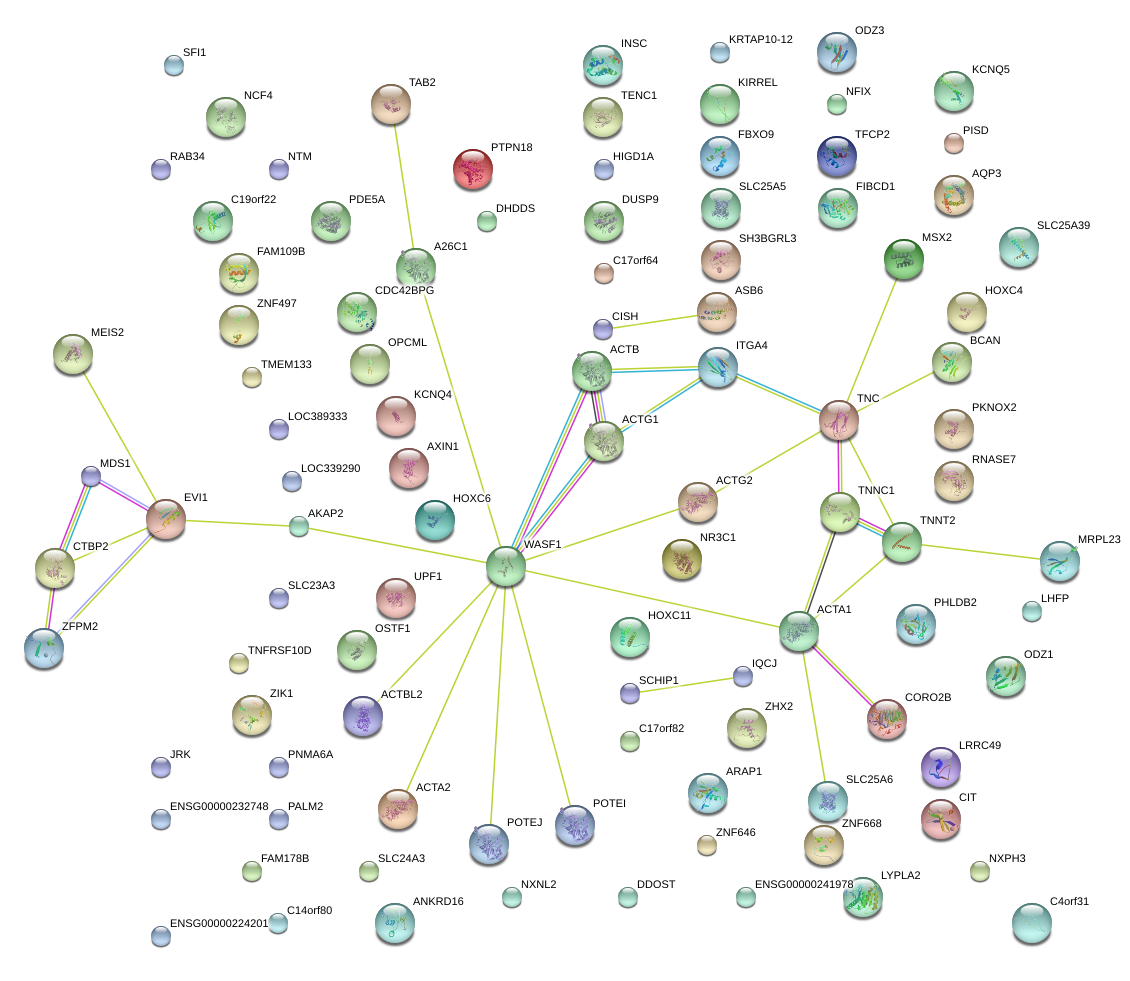
|
chr10_-_126839546
|
2.429
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr2_+_130846263
|
1.642
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr21_+_44941514
|
1.448
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr22_+_35586950
|
1.423
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr4_-_122213018
|
1.367
|
NM_024574
|
C4orf31
|
chromosome 4 open reading frame 31
|
|
chr2_+_182029863
|
1.333
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr11_+_131286513
|
1.214
|
|
NTM
|
neurotrimin
|
|
chr11_+_1925162
|
1.193
|
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr11_-_64368616
|
1.164
|
NM_017525
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr22_+_40800193
|
1.163
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr3_+_113061296
|
1.159
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr5_+_174084137
|
1.120
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr11_+_1925075
|
1.118
|
NM_021134
|
MRPL23
|
mitochondrial ribosomal protein L23
|
|
chr12_+_51727076
|
1.054
|
NM_198316
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_+_23990228
|
1.044
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr3_+_113061330
|
1.040
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr2_+_182030173
|
1.010
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr9_-_33437527
|
1.007
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr11_+_124539814
|
1.003
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr1_-_29322993
|
1.001
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr6_+_73388713
|
1.000
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr22_+_40800213
|
0.985
|
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr3_+_113061218
|
0.983
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr22_-_30356783
|
0.973
|
NM_014338
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr11_+_124539768
|
0.944
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr13_-_39075304
|
0.919
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr3_+_160964574
|
0.888
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_118725672
|
0.879
|
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr1_+_156229934
|
0.866
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr11_-_72111027
|
0.864
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr9_-_131444098
|
0.863
|
NM_017873
NM_177999
|
ASB6
|
ankyrin repeat and SOCS box containing 6
|
|
chr9_-_116920232
|
0.856
|
NM_002160
|
TNC
|
tenascin C
|
|
chr18_+_5228065
|
0.854
|
|
LOC339290
|
hypothetical LOC339290
|
|
chr14_+_105028614
|
0.832
|
NM_001134875
NM_001134876
|
C14orf80
|
chromosome 14 open reading frame 80
|
|
chr15_+_66658626
|
0.808
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr6_+_149680747
|
0.798
|
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr12_-_49853155
|
0.781
|
NM_001173452
NM_001173453
NM_005653
|
TFCP2
|
transcription factor CP2
|
|
chr8_+_106400322
|
0.776
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr3_-_50624085
|
0.773
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr17_-_24068759
|
0.770
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr3_-_50624114
|
0.768
|
NM_013324
NM_145071
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr9_+_76893232
|
0.767
|
|
OSTF1
|
osteoclast stimulating factor 1
|
|
chr16_+_30993252
|
0.767
|
|
ZNF646
|
zinc finger protein 646
|
|
chrX_+_152562013
|
0.762
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr9_+_90339835
|
0.757
|
NM_001161625
NM_145283
|
NXNL2
|
nucleoredoxin-like 2
|
|
chr20_+_19141282
|
0.753
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr8_-_23077392
|
0.740
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr8_-_143748324
|
0.738
|
|
JRK
|
jerky homolog (mouse)
|
|
chr1_+_26479167
|
0.730
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr4_+_183302105
|
0.728
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr2_+_182030510
|
0.722
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr19_+_18803786
|
0.720
|
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr9_-_132804057
|
0.700
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr17_+_45008473
|
0.690
|
|
NXPH3
|
neurexophilin 3
|
|
chr11_+_15093059
|
0.680
|
NM_001042536
|
INSC
|
inscuteable homolog (Drosophila)
|
|
chr16_-_30993004
|
0.674
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr6_-_110607857
|
0.672
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr8_-_143748391
|
0.670
|
NM_001077527
NM_003724
|
JRK
|
jerky homolog (mouse)
|
|
chr1_+_154878344
|
0.670
|
NM_021948
NM_198427
|
BCAN
|
brevican
|
|
chr6_+_53038181
|
0.658
|
NM_033480
|
FBXO9
|
F-box protein 9
|
|
chr12_+_52653176
|
0.650
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr13_-_39075355
|
0.649
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr16_+_30993223
|
0.642
|
NM_014699
|
ZNF646
|
zinc finger protein 646
|
|
chr19_+_12966970
|
0.641
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr17_+_55854636
|
0.641
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chrX_-_151996613
|
0.640
|
NM_032882
|
PNMA6A
|
paraneoplastic antigen like 6A
|
|
chrX_+_118486390
|
0.638
|
NM_001152
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr9_+_111442888
|
0.638
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chrX_+_118486419
|
0.636
|
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5
|
|
chr10_-_5971787
|
0.636
|
NM_001009941
NM_001009943
NM_019046
|
ANKRD16
|
ankyrin repeat domain 16
|
|
chr8_+_123863224
|
0.632
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr16_-_342399
|
0.627
|
NM_003502
NM_181050
|
AXIN1
|
axin 1
|
|
chr15_-_35177664
|
0.623
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_97016027
|
0.621
|
NM_001122646
|
FAM178B
|
family with sequence similarity 178, member B
|
|
chr5_-_138758783
|
0.621
|
NM_001161546
|
LOC389333
|
hypothetical protein LOC389333
|
|
chr12_+_52696908
|
0.620
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr1_-_20860443
|
0.618
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr1_-_20860483
|
0.618
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr15_+_68971835
|
0.617
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr16_-_30992574
|
0.615
|
|
ZNF668
|
zinc finger protein 668
|
|
chr7_-_5535349
|
0.614
|
|
ACTB
|
actin, beta
|
|
chr9_-_132803809
|
0.613
|
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr7_-_5535726
|
0.613
|
|
ACTB
|
actin, beta
|
|
chr5_-_142763418
|
0.610
|
NM_000176
NM_001020825
NM_001024094
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_-_20860524
|
0.609
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr1_-_20860468
|
0.608
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr19_-_63565890
|
0.607
|
NM_198458
|
ZNF497
|
zinc finger protein 497
|
|
chr17_+_56843893
|
0.605
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr4_-_120767777
|
0.601
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr14_+_20580224
|
0.599
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr1_-_199613427
|
0.598
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr17_-_39757725
|
0.596
|
NM_001143780
NM_016016
|
SLC25A39
|
solute carrier family 25, member 39
|
|
chr3_-_170864111
|
0.595
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr1_-_199613406
|
0.595
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr1_+_154351200
|
0.589
|
|
LMNA
|
lamin A/C
|
|
chr9_+_114552871
|
0.589
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr1_-_40009513
|
0.587
|
NM_022120
|
OXCT2
|
3-oxoacid CoA transferase 2
|
|
chr20_-_55719946
|
0.587
|
NM_199171
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr9_+_108665168
|
0.586
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr1_-_20860534
|
0.586
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr5_-_131854325
|
0.584
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr1_+_12046020
|
0.579
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr1_-_12599934
|
0.579
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_-_42274160
|
0.577
|
|
|
|
|
chr19_+_18803742
|
0.576
|
NM_002911
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast)
|
|
chr11_+_9362775
|
0.573
|
|
IPO7
|
importin 7
|
|
chr1_-_108544486
|
0.566
|
NM_013386
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24
|
|
chr1_+_202308814
|
0.564
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr11_+_19095215
|
0.561
|
NM_001001483
NM_019028
|
ZDHHC13
|
zinc finger, DHHC-type containing 13
|
|
chr22_-_28114568
|
0.560
|
NM_001127
NM_001166019
NM_145730
|
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit
|
|
chr1_-_108544416
|
0.559
|
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24
|
|
chr13_-_39075200
|
0.555
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr10_-_134449467
|
0.552
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr1_-_20860454
|
0.550
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr15_+_66357092
|
0.546
|
NM_015322
|
FEM1B
|
fem-1 homolog b (C. elegans)
|
|
chr11_-_103540275
|
0.546
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr12_-_109372481
|
0.545
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr6_-_72187168
|
0.543
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr4_+_156900053
|
0.542
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr15_+_72695248
|
0.538
|
|
CLK3
|
CDC-like kinase 3
|
|
chr12_-_109372462
|
0.538
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr6_+_73388291
|
0.537
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr12_-_46438447
|
0.536
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_168899936
|
0.536
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr11_+_9362699
|
0.534
|
NM_006391
|
IPO7
|
importin 7
|
|
chr4_+_6627816
|
0.533
|
|
MAN2B2
|
mannosidase, alpha, class 2B, member 2
|
|
chr11_-_64266916
|
0.532
|
|
|
|
|
chr2_+_149895274
|
0.532
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr19_+_10843369
|
0.531
|
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr10_+_52504239
|
0.528
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr2_-_27147995
|
0.522
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr3_-_53055078
|
0.521
|
NM_001005158
NM_016329
|
SFMBT1
|
Scm-like with four mbt domains 1
|
|
chr11_-_62363551
|
0.521
|
|
WDR74
|
WD repeat domain 74
|
|
chr11_-_64267037
|
0.520
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr4_+_184663141
|
0.519
|
NM_001564
|
ING2
|
inhibitor of growth family, member 2
|
|
chr17_-_39619549
|
0.515
|
|
C17orf65
|
chromosome 17 open reading frame 65
|
|
chr7_+_150387578
|
0.514
|
NM_003040
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr11_-_9292581
|
0.514
|
|
TMEM41B
|
transmembrane protein 41B
|
|
chr17_+_45008175
|
0.513
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr1_-_20860500
|
0.513
|
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
|
|
chr14_+_103674812
|
0.512
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr3_-_124650081
|
0.511
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr2_-_201082978
|
0.508
|
NM_152387
|
KCTD18
|
potassium channel tetramerisation domain containing 18
|
|
chr11_-_63749916
|
0.508
|
NM_001160393
|
TRPT1
|
tRNA phosphotransferase 1
|
|
chr16_+_3447957
|
0.507
|
NM_001083600
NM_024845
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr1_-_118529303
|
0.506
|
NM_206996
|
SPAG17
|
sperm associated antigen 17
|
|
chr14_+_55654900
|
0.506
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr4_-_122063118
|
0.504
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr11_-_6451561
|
0.504
|
NM_006458
NM_033278
|
TRIM3
|
tripartite motif containing 3
|
|
chr9_-_131845277
|
0.502
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr6_+_137185376
|
0.501
|
NM_000288
|
PEX7
|
peroxisomal biogenesis factor 7
|
|
chr19_-_54646837
|
0.500
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr8_+_22513043
|
0.499
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr7_-_134506070
|
0.499
|
NM_024033
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr1_+_44230011
|
0.499
|
|
CCDC24
|
coiled-coil domain containing 24
|
|
chr11_-_103540233
|
0.499
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr11_+_64635870
|
0.496
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr12_-_109372489
|
0.496
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr22_-_49025499
|
0.494
|
NM_020461
|
TUBGCP6
|
tubulin, gamma complex associated protein 6
|
|
chr9_-_76757555
|
0.491
|
NM_017998
|
C9orf40
|
chromosome 9 open reading frame 40
|
|
chr1_+_26479208
|
0.487
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr6_+_137185441
|
0.487
|
|
PEX7
|
peroxisomal biogenesis factor 7
|
|
chr8_+_121206478
|
0.485
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr3_+_44729138
|
0.484
|
NM_001134440
NM_001134441
NM_001134442
NM_033210
|
ZNF502
|
zinc finger protein 502
|
|
chr16_+_639278
|
0.482
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr1_+_26671488
|
0.482
|
NM_005517
|
HMGN2
|
high-mobility group nucleosomal binding domain 2
|
|
chr15_+_97010219
|
0.482
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr10_-_134605978
|
0.481
|
NM_173572
|
C10orf93
|
chromosome 10 open reading frame 93
|
|
chr22_+_23753940
|
0.481
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr12_-_109372517
|
0.481
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr17_-_39757660
|
0.477
|
|
SLC25A39
|
solute carrier family 25, member 39
|
|
chr17_+_35075124
|
0.477
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr1_+_6537924
|
0.474
|
NM_138697
NM_177540
|
TAS1R1
|
taste receptor, type 1, member 1
|
|
chr22_-_43987010
|
0.474
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr12_-_55920741
|
0.472
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr1_+_152014391
|
0.470
|
NM_024330
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chr11_-_64247230
|
0.470
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr16_+_84204424
|
0.469
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr1_+_153241665
|
0.469
|
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr3_-_120236118
|
0.469
|
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr1_+_112852892
|
0.466
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr6_-_32184975
|
0.465
|
NM_019105
|
TNXB
|
tenascin XB
|
|
chr4_+_6627801
|
0.464
|
NM_015274
|
MAN2B2
|
mannosidase, alpha, class 2B, member 2
|
|
chr4_-_84250019
|
0.464
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr20_+_55359551
|
0.464
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chrX_+_69589081
|
0.463
|
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr19_-_51608665
|
0.462
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr3_+_51964369
|
0.462
|
NM_080865
|
GPR62
|
G protein-coupled receptor 62
|
|
chr8_-_146023228
|
0.460
|
|
LOC100130027
|
hypothetical LOC100130027
|
|
chr10_-_76829857
|
0.460
|
|
ZNF503
|
zinc finger protein 503
|
|
chr22_-_20637188
|
0.460
|
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr17_-_23721351
|
0.459
|
|
VTN
|
vitronectin
|
|
chrX_-_99551926
|
0.459
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr14_+_55654607
|
0.457
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chrX_+_133993984
|
0.456
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr7_-_100331417
|
0.456
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr10_+_44816278
|
0.455
|
NM_006963
|
ZNF22
|
zinc finger protein 22 (KOX 15)
|
|
chr1_+_179148935
|
0.454
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr22_+_36286350
|
0.454
|
NM_152243
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr20_+_55359706
|
0.453
|
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|