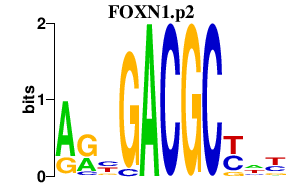
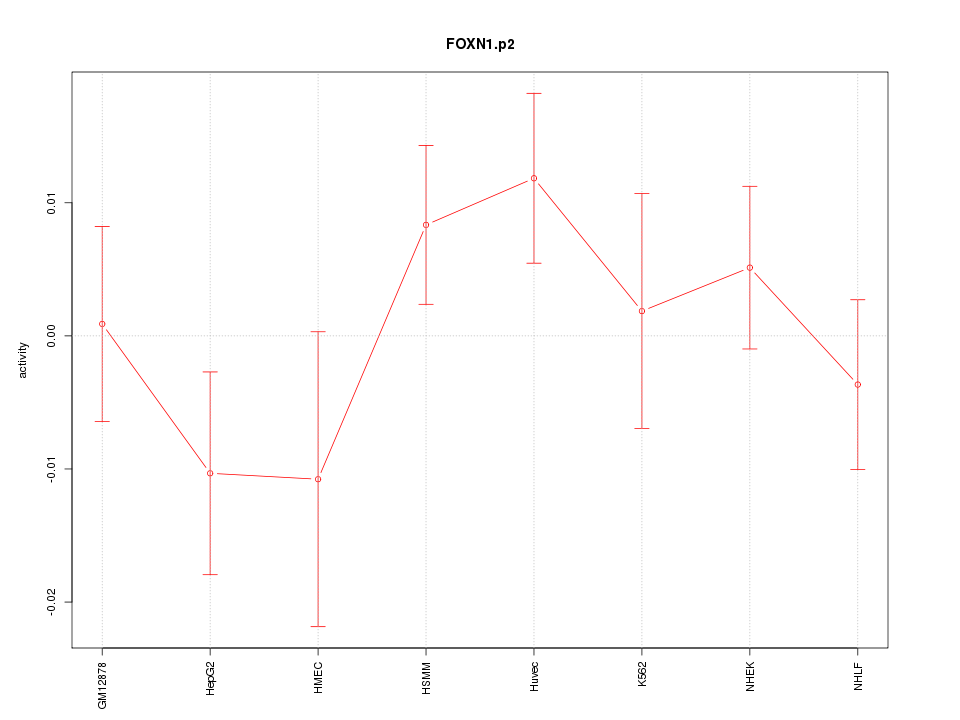
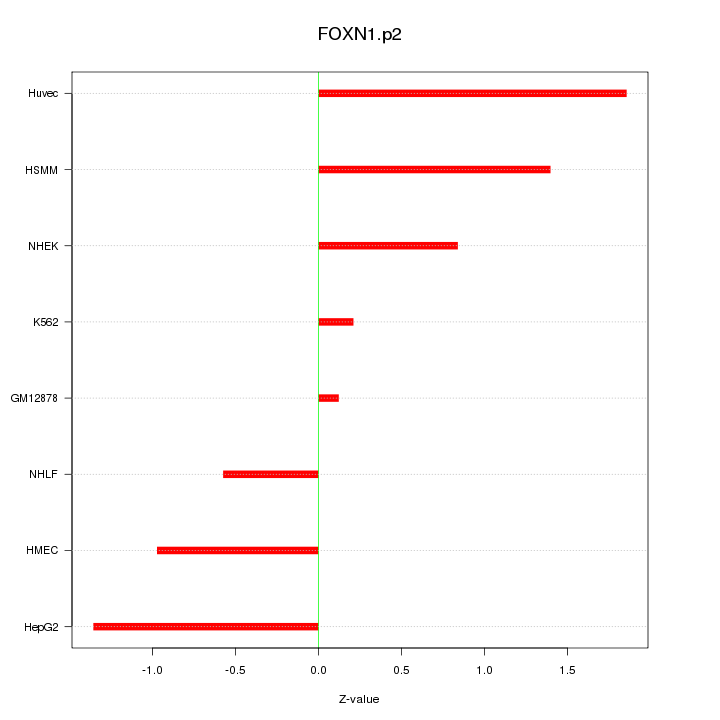
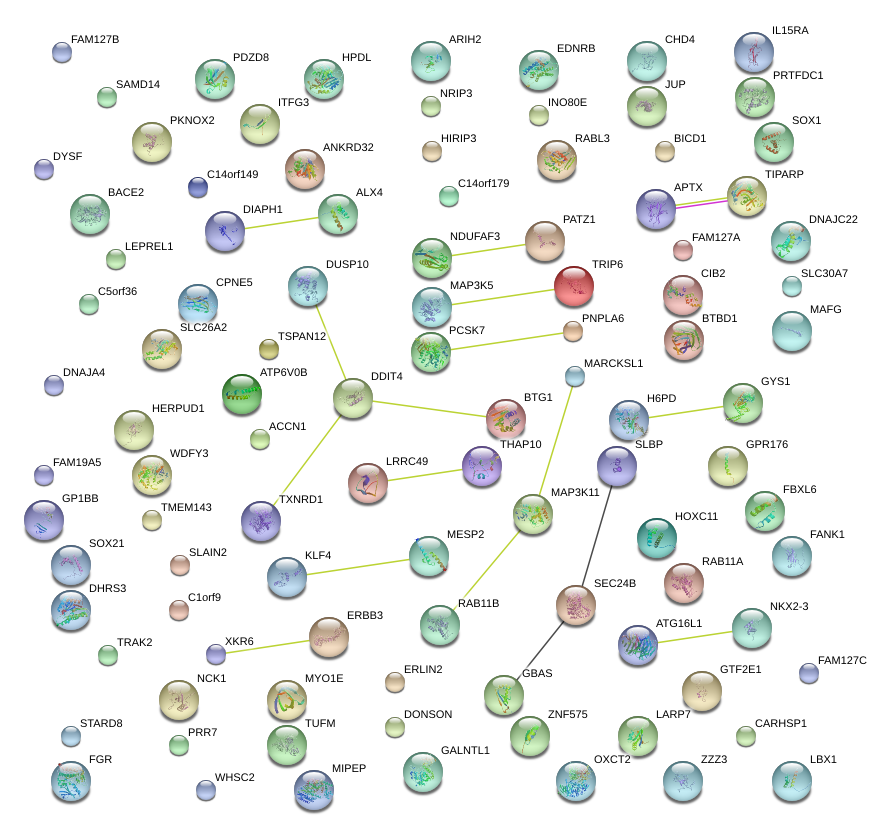
|
chr7_-_120285364
|
1.249
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr7_-_120285245
|
1.063
|
|
TSPAN12
|
tetraspanin 12
|
|
chr2_+_71534260
|
0.724
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr15_+_76343540
|
0.704
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_+_68971835
|
0.614
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr15_-_68971665
|
0.610
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr6_-_36915195
|
0.598
|
NM_020939
|
CPNE5
|
copine V
|
|
chr4_-_1980473
|
0.574
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr13_-_77390907
|
0.568
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr8_-_11096257
|
0.537
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr10_-_102978678
|
0.524
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr4_-_86106567
|
0.522
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr14_+_75521806
|
0.482
|
NM_001102564
NM_052873
|
C14orf179
|
chromosome 14 open reading frame 179
|
|
chr4_-_1980500
|
0.480
|
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr12_+_48026955
|
0.479
|
|
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr10_+_101282679
|
0.464
|
NM_145285
|
NKX2-3
|
NK2 transcription factor related, locus 3 (Drosophila)
|
|
chr5_-_140978751
|
0.453
|
NM_001079812
NM_005219
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr3_+_157876051
|
0.453
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr5_+_93980108
|
0.450
|
NM_032290
|
ANKRD32
|
ankyrin repeat domain 32
|
|
chr4_+_110574274
|
0.445
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr5_+_176806401
|
0.444
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr12_-_91063441
|
0.436
|
NM_001731
|
BTG1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr4_-_1980756
|
0.435
|
NM_005663
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr15_-_76210611
|
0.435
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr1_-_27834210
|
0.429
|
NM_005248
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr3_-_121944052
|
0.424
|
NM_173825
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr9_-_109290107
|
0.420
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr5_-_140978744
|
0.420
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr15_+_53488014
|
0.418
|
NM_001198784
|
FLJ27352
|
hypothetical LOC145788
|
|
chr6_-_137155222
|
0.414
|
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr13_-_94162249
|
0.412
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr3_+_138063764
|
0.411
|
|
NCK1
|
NCK adaptor protein 1
|
|
chr17_-_28644118
|
0.407
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr5_+_149320480
|
0.405
|
NM_000112
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr12_+_54760211
|
0.400
|
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr4_+_110574376
|
0.394
|
NM_001042734
NM_006323
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr22_+_47263919
|
0.391
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr5_-_140978747
|
0.387
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr4_-_110574363
|
0.382
|
|
1/2-SBSRNA4
|
lncRNA_BC009800
|
|
chr2_-_39040981
|
0.380
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr10_+_73703682
|
0.380
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr19_-_54188332
|
0.366
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr21_-_33882807
|
0.365
|
NM_017613
|
DONSON
|
downstream neighbor of SON
|
|
chr22_-_30071565
|
0.361
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr19_-_54188376
|
0.361
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr16_-_28764928
|
0.359
|
|
TUFM
|
Tu translation elongation factor, mitochondrial
|
|
chr17_-_37196463
|
0.357
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr12_+_48027307
|
0.356
|
NM_024902
|
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr12_+_32150922
|
0.355
|
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr15_-_57452357
|
0.354
|
|
MYO1E
|
myosin IE
|
|
chr15_-_57452345
|
0.349
|
|
MYO1E
|
myosin IE
|
|
chr19_-_54188378
|
0.348
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr15_-_57452361
|
0.346
|
NM_004998
|
MYO1E
|
myosin IE
|
|
chr19_-_54188181
|
0.344
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr1_+_9217444
|
0.343
|
NM_004285
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr17_-_77478835
|
0.343
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr16_+_55523248
|
0.343
|
NM_001010989
NM_001010990
NM_014685
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1
|
|
chr3_-_121944040
|
0.340
|
|
|
|
|
chr6_-_137155295
|
0.339
|
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr1_+_101134276
|
0.339
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr16_-_8870343
|
0.337
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr15_-_38000379
|
0.336
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chrX_-_134013644
|
0.335
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr5_-_140978664
|
0.335
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr4_+_110574420
|
0.335
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr12_+_103133631
|
0.334
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr6_-_137155317
|
0.334
|
NM_005923
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr12_+_32151351
|
0.333
|
NM_001003398
NM_001714
|
BICD1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr10_-_25281510
|
0.332
|
NM_020200
|
PRTFDC1
|
phosphoribosyl transferase domain containing 1
|
|
chr3_-_191321367
|
0.331
|
|
LEPREL1
|
leprecan-like 1
|
|
chr16_+_29915003
|
0.330
|
NM_173618
|
INO80E
|
INO80 complex subunit E
|
|
chr22_+_18091065
|
0.328
|
NM_000407
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide
|
|
chr5_-_157011665
|
0.327
|
|
|
|
|
chr3_-_191321601
|
0.326
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr1_-_77921681
|
0.324
|
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr19_+_7505723
|
0.323
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr10_-_119124727
|
0.321
|
NM_173791
|
PDZD8
|
PDZ domain containing 8
|
|
chr7_+_100302885
|
0.319
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr1_-_40009513
|
0.319
|
NM_022120
|
OXCT2
|
3-oxoacid CoA transferase 2
|
|
chr19_+_7505590
|
0.315
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr12_-_6585785
|
0.314
|
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr3_+_138063697
|
0.313
|
NM_006153
|
NCK1
|
NCK adaptor protein 1
|
|
chr15_-_81527109
|
0.312
|
NM_001011885
NM_025238
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr5_+_149320539
|
0.310
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr1_+_9217384
|
0.309
|
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr11_-_116607968
|
0.309
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr4_+_113777568
|
0.307
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr16_-_29914816
|
0.306
|
NM_003609
|
HIRIP3
|
HIRA interacting protein 3
|
|
chr3_+_121944247
|
0.305
|
NM_005513
|
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa
|
|
chr6_-_137155414
|
0.305
|
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr5_-_93979932
|
0.301
|
NM_001145678
NM_173665
|
KIAA0825
|
KIAA0825
|
|
chr19_-_53558961
|
0.301
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr11_-_44288188
|
0.299
|
NM_021926
|
ALX4
|
ALX homeobox 4
|
|
chr1_-_12599934
|
0.298
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr8_+_37713313
|
0.297
|
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr15_+_88120592
|
0.294
|
NM_001039958
|
MESP2
|
mesoderm posterior 2 homolog (mouse)
|
|
chr3_+_49034050
|
0.293
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr10_+_127575097
|
0.292
|
NM_145235
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1
|
|
chr11_-_65139277
|
0.291
|
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr1_-_219982016
|
0.290
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr10_-_6059460
|
0.290
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr8_+_37713295
|
0.288
|
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr12_+_52653176
|
0.287
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr1_+_101134259
|
0.287
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr1_+_45565126
|
0.287
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr11_+_124539814
|
0.287
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr17_-_60208083
|
0.284
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr8_+_37713236
|
0.284
|
NM_001003791
NM_007175
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr1_-_32574185
|
0.283
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr16_+_29915063
|
0.282
|
|
INO80E
|
INO80 complex subunit E
|
|
chr5_-_36912487
|
0.282
|
|
LOC646719
|
hypothetical LOC646719
|
|
chr21_+_41461962
|
0.279
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_-_219982049
|
0.278
|
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr1_+_44213182
|
0.277
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr15_+_63948823
|
0.276
|
NM_004663
|
RAB11A
|
RAB11A, member RAS oncogene family
|
|
chr1_+_44213200
|
0.276
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr19_+_48729127
|
0.275
|
NM_174945
|
ZNF575
|
zinc finger protein 575
|
|
chr6_-_137154859
|
0.275
|
|
|
|
|
chr4_-_1683712
|
0.274
|
NM_006527
|
SLBP
|
stem-loop binding protein
|
|
chr16_+_224834
|
0.272
|
|
ITFG3
|
integrin alpha FG-GAP repeat containing 3
|
|
chr9_-_32991518
|
0.270
|
NM_001195249
NM_001195254
NM_001195248
NM_001195250
NM_001195251
NM_001195252
NM_175069
NM_175073
|
APTX
|
aprataxin
|
|
chr4_+_48038095
|
0.269
|
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr17_+_77478858
|
0.269
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr15_-_81527002
|
0.269
|
|
BTBD1
|
BTB (POZ) domain containing 1
|
|
chr1_+_101134237
|
0.268
|
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr2_+_233825056
|
0.265
|
|
ATG16L1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae)
|
|
chr17_-_45562112
|
0.265
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr3_+_48931248
|
0.264
|
|
ARIH2
|
ariadne homolog 2 (Drosophila)
|
|
chr11_-_8982107
|
0.263
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr7_+_55999559
|
0.261
|
NM_001483
|
GBAS
|
glioblastoma amplified sequence
|
|
chrX_+_67830215
|
0.261
|
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr15_-_37999717
|
0.260
|
|
GPR176
|
G protein-coupled receptor 176
|
|
chr2_+_233825084
|
0.260
|
|
ATG16L1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae)
|
|
chr7_-_106088565
|
0.260
|
NM_175884
|
FLJ36031
|
hypothetical protein FLJ36031
|
|
chr14_+_68796433
|
0.260
|
NM_001168368
NM_020692
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr1_+_170768635
|
0.259
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chrX_+_67830204
|
0.259
|
NM_001142504
|
STARD8
|
StAR-related lipid transfer (START) domain containing 8
|
|
chr2_+_233824955
|
0.259
|
NM_001190266
NM_001190267
NM_017974
NM_030803
NM_198890
|
ATG16L1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae)
|
|
chr14_-_59020795
|
0.258
|
NM_144581
|
C14orf149
|
chromosome 14 open reading frame 149
|
|
chr1_+_101134177
|
0.257
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr2_-_202024317
|
0.256
|
|
TRAK2
|
trafficking protein, kinesin binding 2
|
|
chr13_-_23361430
|
0.254
|
NM_005932
|
MIPEP
|
mitochondrial intermediate peptidase
|
|
chr9_+_81377289
|
0.253
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr5_-_100266769
|
0.252
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr5_-_142763252
|
0.250
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr12_+_63849606
|
0.250
|
NM_001167614
NM_014319
|
LEMD3
|
LEM domain containing 3
|
|
chr11_+_124539768
|
0.250
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chrX_+_67635593
|
0.248
|
|
YIPF6
|
Yip1 domain family, member 6
|
|
chr10_+_112394094
|
0.248
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr16_+_224799
|
0.248
|
NM_032039
|
ITFG3
|
integrin alpha FG-GAP repeat containing 3
|
|
chr1_-_1233109
|
0.248
|
NM_030649
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3
|
|
chr4_+_57066131
|
0.247
|
NM_206919
|
ARL9
|
ADP-ribosylation factor-like 9
|
|
chr15_-_53487748
|
0.247
|
NM_004748
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr21_-_39607415
|
0.247
|
NM_001007246
NM_018963
NM_033656
|
BRWD1
|
bromodomain and WD repeat domain containing 1
|
|
chr8_+_37713329
|
0.245
|
|
ERLIN2
|
ER lipid raft associated 2
|
|
chr6_+_126112417
|
0.244
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr1_+_44213229
|
0.244
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr9_+_127550298
|
0.244
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr18_+_9698187
|
0.242
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr5_+_36912595
|
0.241
|
NM_015384
NM_133433
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr7_-_44854222
|
0.240
|
NM_012412
NM_138635
NM_201436
NM_201516
NM_201517
|
H2AFV
|
H2A histone family, member V
|
|
chr13_+_48448950
|
0.239
|
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_-_2250967
|
0.238
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr5_+_151131704
|
0.233
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr7_+_138566770
|
0.233
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr12_-_50502474
|
0.233
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chr12_+_4889309
|
0.233
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr12_+_54760153
|
0.233
|
NM_001005915
NM_001982
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr2_+_232534535
|
0.233
|
NM_152383
|
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2
|
|
chr5_+_149320630
|
0.233
|
|
SLC26A2
|
solute carrier family 26 (sulfate transporter), member 2
|
|
chr5_+_141328915
|
0.233
|
NM_183401
|
RNF14
|
ring finger protein 14
|
|
chr10_-_52053617
|
0.232
|
NM_147156
|
SGMS1
|
sphingomyelin synthase 1
|
|
chr19_-_11500912
|
0.231
|
NM_001142464
NM_001142465
NM_016581
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr1_+_170768914
|
0.231
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr1_+_44213210
|
0.228
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr11_+_9362775
|
0.226
|
|
IPO7
|
importin 7
|
|
chr1_-_209818675
|
0.225
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr15_+_45796802
|
0.224
|
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr13_+_20770248
|
0.223
|
|
LOC650794
|
hypothetical LOC650794
|
|
chr18_+_17076166
|
0.223
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr1_-_219982081
|
0.223
|
NM_007207
NM_144729
|
DUSP10
|
dual specificity phosphatase 10
|
|
chr15_+_39008822
|
0.222
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr5_+_151131676
|
0.222
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_+_179180447
|
0.222
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr6_+_126112506
|
0.222
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr5_-_159478967
|
0.221
|
NM_001130864
NM_052927
|
PWWP2A
|
PWWP domain containing 2A
|
|
chr1_-_29380969
|
0.221
|
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr1_-_209818576
|
0.220
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr17_-_37586716
|
0.220
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr5_+_179180517
|
0.219
|
|
SQSTM1
|
sequestosome 1
|
|
chr2_+_220007943
|
0.219
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr3_-_100103142
|
0.218
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr22_+_28806452
|
0.218
|
NM_152510
|
HORMAD2
|
HORMA domain containing 2
|
|
chr13_-_77391525
|
0.216
|
|
|
|
|
chr16_+_19087035
|
0.216
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr10_-_30065760
|
0.215
|
|
SVIL
|
supervillin
|
|
chr17_+_45940743
|
0.215
|
NM_032133
|
MYCBPAP
|
MYCBP associated protein
|
|
chr11_-_67029349
|
0.214
|
NM_001130848
NM_004910
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr1_-_29380945
|
0.214
|
|
SRSF4
|
serine/arginine-rich splicing factor 4
|
|
chr15_+_81567327
|
0.214
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|