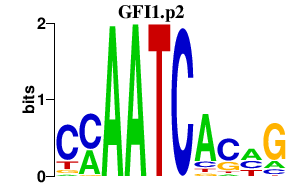
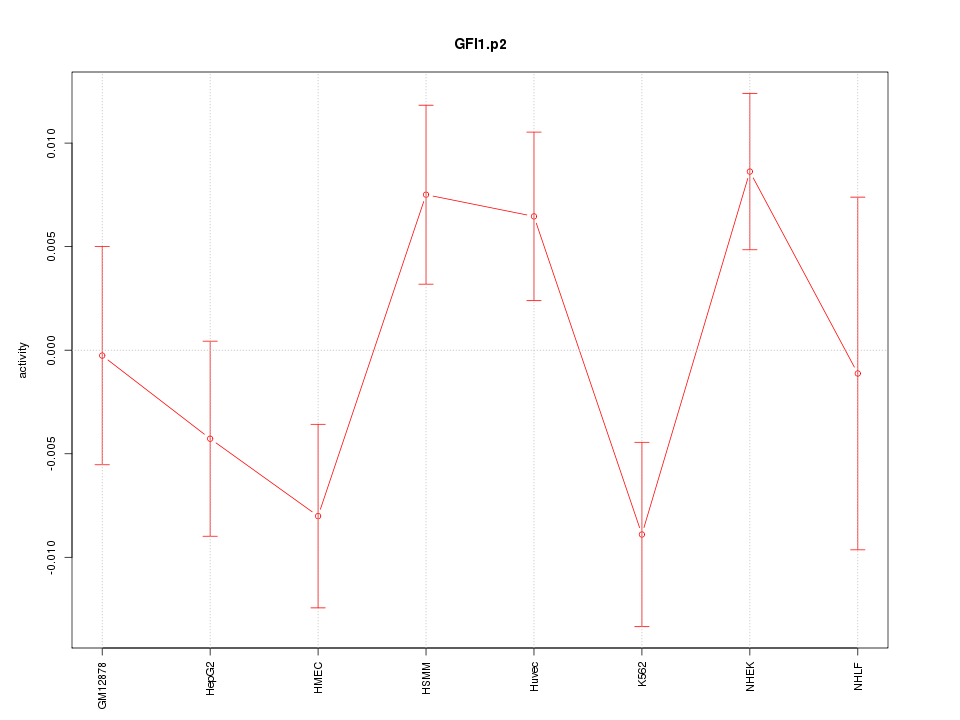
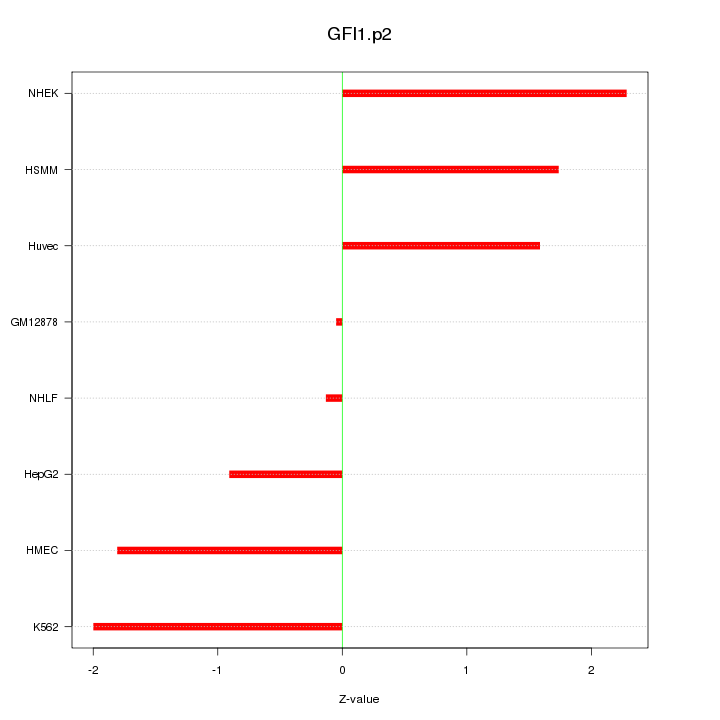
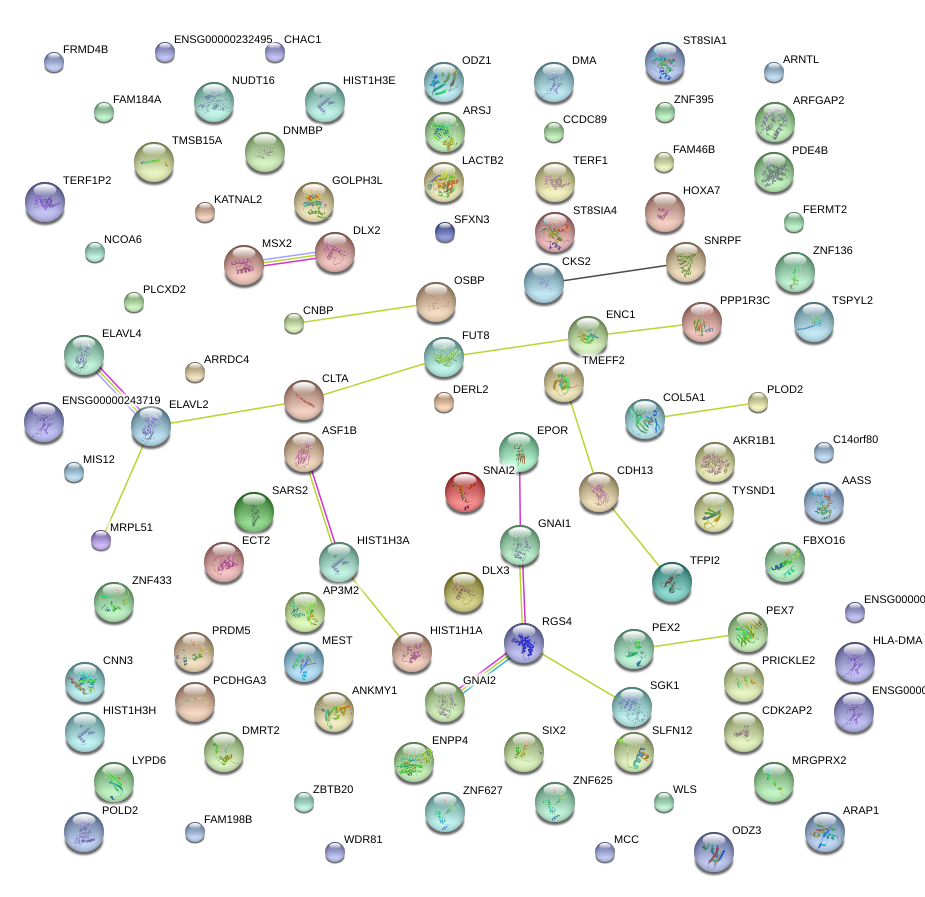
|
chr17_+_1566566
|
1.129
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr14_+_105028614
|
1.039
|
NM_001134875
NM_001134876
|
C14orf80
|
chromosome 14 open reading frame 80
|
|
chr4_+_183302105
|
1.030
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr17_-_1566223
|
1.014
|
|
C17orf91
|
chromosome 17 open reading frame 91
|
|
chr1_+_161305558
|
0.997
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_161305188
|
0.975
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr7_-_27162688
|
0.963
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr5_+_174084137
|
0.936
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr3_+_132583204
|
0.919
|
NM_001171905
NM_001171906
NM_152395
|
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr3_+_121296290
|
0.903
|
|
|
|
|
chr2_+_149894744
|
0.856
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr4_-_115120251
|
0.854
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr10_-_71576458
|
0.850
|
NM_001040273
NM_173555
|
TYSND1
|
trypsin domain containing 1
|
|
chr9_+_136673534
|
0.840
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr11_-_47154955
|
0.840
|
NM_032389
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr19_-_11352850
|
0.837
|
|
EPOR
|
erythropoietin receptor
|
|
chr3_+_112876212
|
0.819
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr8_-_78074851
|
0.809
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr11_-_47154996
|
0.804
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr9_-_23816062
|
0.798
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_-_33028790
|
0.796
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr19_-_14108267
|
0.793
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr8_+_42129620
|
0.771
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr14_+_64948332
|
0.757
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr1_-_148936123
|
0.757
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr17_-_5330204
|
0.744
|
NM_016041
|
DERL2
|
Der1-like domain family, member 2
|
|
chr19_-_14108400
|
0.742
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr6_-_26125939
|
0.733
|
NM_005325
|
HIST1H1A
|
histone cluster 1, H1a
|
|
chr15_+_39032927
|
0.713
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr3_-_116348780
|
0.710
|
NM_015642
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr11_-_85074848
|
0.692
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chr5_-_73973004
|
0.685
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr7_-_133794344
|
0.659
|
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase)
|
|
chr17_-_43328141
|
0.652
|
|
|
|
|
chr7_-_133794404
|
0.638
|
NM_001628
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase)
|
|
chr1_+_66570683
|
0.631
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr10_+_102782156
|
0.630
|
|
SFXN3
|
sideroflexin 3
|
|
chr11_-_19038803
|
0.624
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr13_-_77391525
|
0.624
|
|
|
|
|
chrX_-_101658297
|
0.623
|
NM_021992
|
TMSB15A
TMSB15B
|
thymosin beta 15a
thymosin beta 15B
|
|
chr3_-_64186151
|
0.622
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_+_66570363
|
0.616
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr9_+_1040600
|
0.616
|
NM_181872
|
DMRT2
|
doublesex and mab-3 related transcription factor 2
|
|
chr5_-_157011665
|
0.581
|
|
|
|
|
chr6_+_26333361
|
0.580
|
NM_003532
|
HIST1H3E
|
histone cluster 1, H3e
|
|
chr10_-_71576415
|
0.576
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr8_-_28403652
|
0.571
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr9_+_36180885
|
0.564
|
|
CLTA
|
clathrin, light chain A
|
|
chr1_-_68470585
|
0.556
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr9_+_36180852
|
0.556
|
NM_001076677
NM_001184760
NM_001184761
NM_001184762
NM_001833
NM_007096
|
CLTA
|
clathrin, light chain A
|
|
chr3_+_173951163
|
0.555
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr5_-_92982927
|
0.546
|
|
|
|
|
chr10_+_102782151
|
0.544
|
|
SFXN3
|
sideroflexin 3
|
|
chr7_+_129918408
|
0.543
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr9_+_1040367
|
0.542
|
|
DMRT2
|
doublesex and mab-3 related transcription factor 2
|
|
chr9_+_1040333
|
0.536
|
NM_006557
NM_001130865
|
DMRT2
|
doublesex and mab-3 related transcription factor 2
|
|
chr9_+_36180927
|
0.533
|
|
CLTA
|
clathrin, light chain A
|
|
chr9_+_136673464
|
0.532
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr4_-_159313613
|
0.530
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr11_-_72111027
|
0.528
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr2_+_149895274
|
0.514
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr4_-_122063118
|
0.514
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr10_-_101680645
|
0.512
|
|
DNMBP
|
dynamin binding protein
|
|
chr10_-_71576335
|
0.510
|
|
|
|
|
chr9_+_36180936
|
0.508
|
|
CLTA
|
clathrin, light chain A
|
|
chr6_-_119512035
|
0.504
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr12_+_94776901
|
0.501
|
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr5_+_140772691
|
0.499
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr3_-_130385454
|
0.483
|
NM_001127192
NM_001127193
NM_001127194
NM_001127195
NM_001127196
NM_003418
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr6_+_26128656
|
0.481
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr9_+_36180956
|
0.480
|
|
CLTA
|
clathrin, light chain A
|
|
chr12_+_94776836
|
0.479
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr17_+_5330439
|
0.479
|
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr8_-_71743908
|
0.477
|
|
LACTB2
|
lactamase, beta 2
|
|
chr7_-_121571472
|
0.476
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr5_-_100266769
|
0.468
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr10_-_93382735
|
0.465
|
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr8_+_74083615
|
0.456
|
NM_003218
NM_017489
|
TERF1
|
telomeric repeat binding factor (NIMA-interacting) 1
|
|
chr6_-_134540666
|
0.454
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_6473207
|
0.454
|
|
MRPL51
|
mitochondrial ribosomal protein L51
|
|
chr3_-_69517913
|
0.449
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr6_+_137185441
|
0.448
|
|
PEX7
|
peroxisomal biogenesis factor 7
|
|
chr18_+_42780784
|
0.448
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr6_+_137185376
|
0.445
|
NM_000288
|
PEX7
|
peroxisomal biogenesis factor 7
|
|
chr11_-_59140192
|
0.444
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr11_+_13255836
|
0.438
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr7_-_93357881
|
0.437
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr3_-_147361626
|
0.437
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr3_+_50248652
|
0.437
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr7_-_44129661
|
0.433
|
|
POLD2
|
polymerase (DNA directed), delta 2, regulatory subunit 50kDa
|
|
chr16_+_81218074
|
0.431
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr12_+_46878406
|
0.430
|
|
DKFZP779L1853
|
hypothetical LOC643162
|
|
chr3_+_50248579
|
0.429
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chrX_+_53128207
|
0.429
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr3_+_50248639
|
0.424
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr9_+_91115924
|
0.423
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr12_-_22378833
|
0.422
|
NM_003034
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
|
|
chr2_-_192767494
|
0.419
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr2_-_241146040
|
0.412
|
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr3_+_50248649
|
0.412
|
NM_002070
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr5_-_112658456
|
0.411
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr10_-_93382779
|
0.410
|
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr3_-_130385444
|
0.404
|
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr19_-_44112920
|
0.402
|
NM_001145901
NM_017827
|
SARS2
|
seryl-tRNA synthetase 2, mitochondrial
|
|
chr19_-_12128528
|
0.402
|
NM_145233
|
ZNF625-ZNF20
ZNF625
|
ZNF625-ZNF20 read-through transcript
zinc finger protein 625
|
|
chr17_-_30783631
|
0.401
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr6_+_46205777
|
0.401
|
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr15_+_96304911
|
0.401
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr2_-_45090025
|
0.397
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr10_-_93382837
|
0.396
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chr8_-_49996353
|
0.394
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr3_+_50248678
|
0.389
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr1_-_95165037
|
0.385
|
|
CNN3
|
calponin 3, acidic
|
|
chr11_-_67032125
|
0.384
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr17_-_45427565
|
0.384
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr1_-_95165089
|
0.384
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr14_-_52487558
|
0.383
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_-_241146077
|
0.380
|
NM_016552
NM_017844
|
ANKMY1
|
ankyrin repeat and MYND domain containing 1
|
|
chr1_-_27211913
|
0.378
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr4_+_95898098
|
0.377
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr1_+_172060563
|
0.376
|
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial
|
|
chr19_+_18164989
|
0.375
|
NM_032683
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2
|
|
chr12_-_55013987
|
0.375
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr1_+_172060440
|
0.373
|
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial
|
|
chr4_-_24590867
|
0.372
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr17_+_59933541
|
0.371
|
NM_138363
|
CCDC45
|
coiled-coil domain containing 45
|
|
chr1_+_172060389
|
0.366
|
NM_018122
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial
|
|
chr5_-_112658200
|
0.365
|
|
MCC
|
mutated in colorectal cancers
|
|
chr14_-_52487386
|
0.364
|
|
FERMT2
|
fermitin family member 2
|
|
chr19_-_52709975
|
0.364
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr20_+_4650499
|
0.363
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr1_-_27212035
|
0.362
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr15_+_98960277
|
0.362
|
NM_024708
NM_198243
|
ASB7
|
ankyrin repeat and SOCS box containing 7
|
|
chr10_+_92621221
|
0.359
|
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr22_-_26645214
|
0.358
|
NM_012399
|
PITPNB
|
phosphatidylinositol transfer protein, beta
|
|
chr4_-_138673014
|
0.358
|
|
PCDH18
|
protocadherin 18
|
|
chr15_-_98699628
|
0.358
|
NM_139057
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17
|
|
chr11_-_40272239
|
0.358
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr19_-_54646837
|
0.356
|
NM_017916
|
PIH1D1
|
PIH1 domain containing 1
|
|
chr10_-_16604009
|
0.355
|
NM_001010908
|
C1QL3
|
complement component 1, q subcomponent-like 3
|
|
chr10_+_102123589
|
0.355
|
|
NCRNA00263
|
non-protein coding RNA 263
|
|
chr3_-_102878301
|
0.355
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr11_+_118710424
|
0.354
|
NM_032015
|
RNF26
|
ring finger protein 26
|
|
chr5_+_140742585
|
0.353
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr8_+_42247954
|
0.352
|
NM_001190722
NM_001556
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr16_+_31391998
|
0.350
|
NM_015927
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr17_+_40089270
|
0.350
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr15_+_96304942
|
0.348
|
|
ARRDC4
|
arrestin domain containing 4
|
|
chr11_-_8849485
|
0.348
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr16_+_55836480
|
0.346
|
NM_012106
|
ARL2BP
|
ADP-ribosylation factor-like 2 binding protein
|
|
chr12_+_27568311
|
0.345
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr8_-_71743973
|
0.341
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr7_-_42243318
|
0.338
|
|
GLI3
|
GLI family zinc finger 3
|
|
chr3_+_185537969
|
0.338
|
NM_144635
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr22_-_26527469
|
0.337
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr3_+_50248749
|
0.335
|
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr22_+_31080871
|
0.334
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr19_+_44113402
|
0.334
|
NM_021107
NM_033363
|
MRPS12
|
mitochondrial ribosomal protein S12
|
|
chr19_-_1993022
|
0.334
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr12_+_53534565
|
0.334
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chr16_+_28798735
|
0.333
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr4_-_83938920
|
0.333
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr1_+_101475149
|
0.332
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_+_59933646
|
0.330
|
|
CCDC45
|
coiled-coil domain containing 45
|
|
chr5_-_88215034
|
0.329
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_179324260
|
0.329
|
NM_016545
|
IER5
|
immediate early response 5
|
|
chr3_+_56566222
|
0.328
|
NM_001012506
NM_001141947
|
CCDC66
|
coiled-coil domain containing 66
|
|
chr14_-_75196911
|
0.327
|
|
C14orf1
|
chromosome 14 open reading frame 1
|
|
chr19_+_20049800
|
0.327
|
NM_007138
|
ZNF90
|
zinc finger protein 90
|
|
chr1_-_7998265
|
0.326
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr8_+_42247991
|
0.326
|
NM_001190720
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr5_+_140835788
|
0.325
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr14_-_103383483
|
0.324
|
NM_015316
|
PPP1R13B
|
protein phosphatase 1, regulatory (inhibitor) subunit 13B
|
|
chr17_-_76827479
|
0.324
|
NM_144679
|
C17orf56
|
chromosome 17 open reading frame 56
|
|
chr8_-_124477639
|
0.323
|
|
ATAD2
|
ATPase family, AAA domain containing 2
|
|
chr1_+_112740378
|
0.322
|
|
|
|
|
chr22_-_35502059
|
0.322
|
NM_001177701
NM_006860
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr6_+_46205659
|
0.321
|
NM_014936
|
ENPP4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative)
|
|
chr9_+_36181015
|
0.321
|
|
CLTA
|
clathrin, light chain A
|
|
chr11_-_55492514
|
0.321
|
NM_001005491
|
OR10AG1
|
olfactory receptor, family 10, subfamily AG, member 1
|
|
chr5_-_92942680
|
0.319
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr3_-_130385362
|
0.319
|
|
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein
|
|
chr12_-_8656705
|
0.318
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chrX_-_103060777
|
0.318
|
|
LOC100128594
|
hypothetical LOC100128594
|
|
chr17_+_43480674
|
0.318
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr4_+_6322459
|
0.317
|
NM_001145853
NM_006005
|
WFS1
|
Wolfram syndrome 1 (wolframin)
|
|
chr4_-_138673078
|
0.316
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr3_-_87122958
|
0.316
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_-_172060399
|
0.316
|
NM_001127181
NM_033319
|
CENPL
|
centromere protein L
|
|
chr14_+_73775927
|
0.315
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr1_+_101474892
|
0.315
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr17_-_57295451
|
0.314
|
NM_032043
|
BRIP1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr12_+_6473519
|
0.312
|
|
NCAPD2
|
non-SMC condensin I complex, subunit D2
|
|
chr17_+_5330588
|
0.312
|
|
MIS12
|
MIS12, MIND kinetochore complex component, homolog (S. pombe)
|
|
chr7_+_87095640
|
0.311
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr1_+_112740380
|
0.310
|
|
LOC100129406
CTTNBP2NL
|
hypothetical protein LOC100129406
CTTNBP2 N-terminal like
|
|
chr10_-_99436889
|
0.310
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr5_+_140835727
|
0.310
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr1_+_169549996
|
0.309
|
NM_002022
|
FMO4
|
flavin containing monooxygenase 4
|
|
chr1_+_160868932
|
0.308
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|