|
chr1_+_239762179
|
2.684
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr13_+_48582389
|
2.238
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr5_+_140227951
|
2.148
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr5_+_140160966
|
2.107
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr9_+_109131814
|
1.948
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr3_+_29297806
|
1.918
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr10_+_91077581
|
1.892
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr14_+_38804226
|
1.861
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr3_-_57301749
|
1.836
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr1_-_240679380
|
1.823
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr14_-_94669547
|
1.823
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr7_+_50106227
|
1.809
|
NM_001161834
|
C7orf72
|
chromosome 7 open reading frame 72
|
|
chrX_-_15593074
|
1.787
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr12_+_67366989
|
1.760
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr9_-_92444886
|
1.746
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr6_+_127940011
|
1.740
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr1_+_150852910
|
1.729
|
NM_178433
|
LCE3B
|
late cornified envelope 3B
|
|
chr17_+_63338500
|
1.705
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr4_+_68107039
|
1.647
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr7_-_14847479
|
1.643
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chrX_-_114158388
|
1.641
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr11_+_65027721
|
1.637
|
|
|
|
|
chr2_-_213723298
|
1.610
|
NM_016260
|
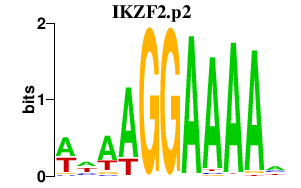
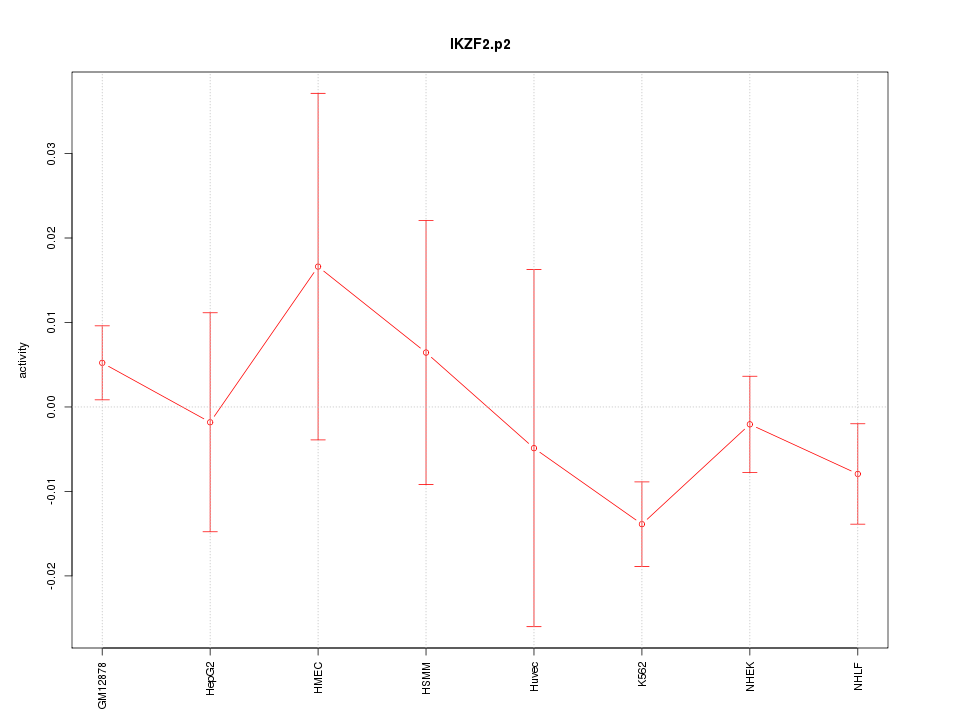
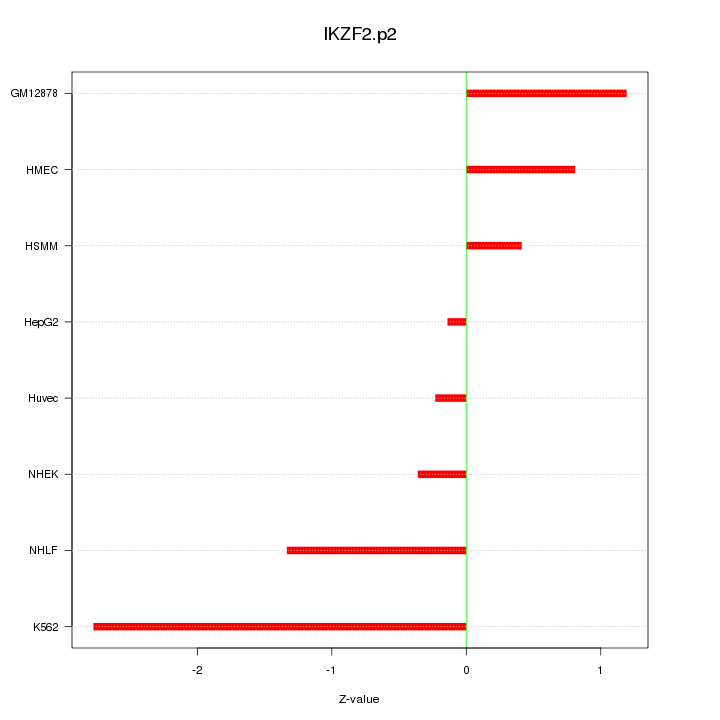
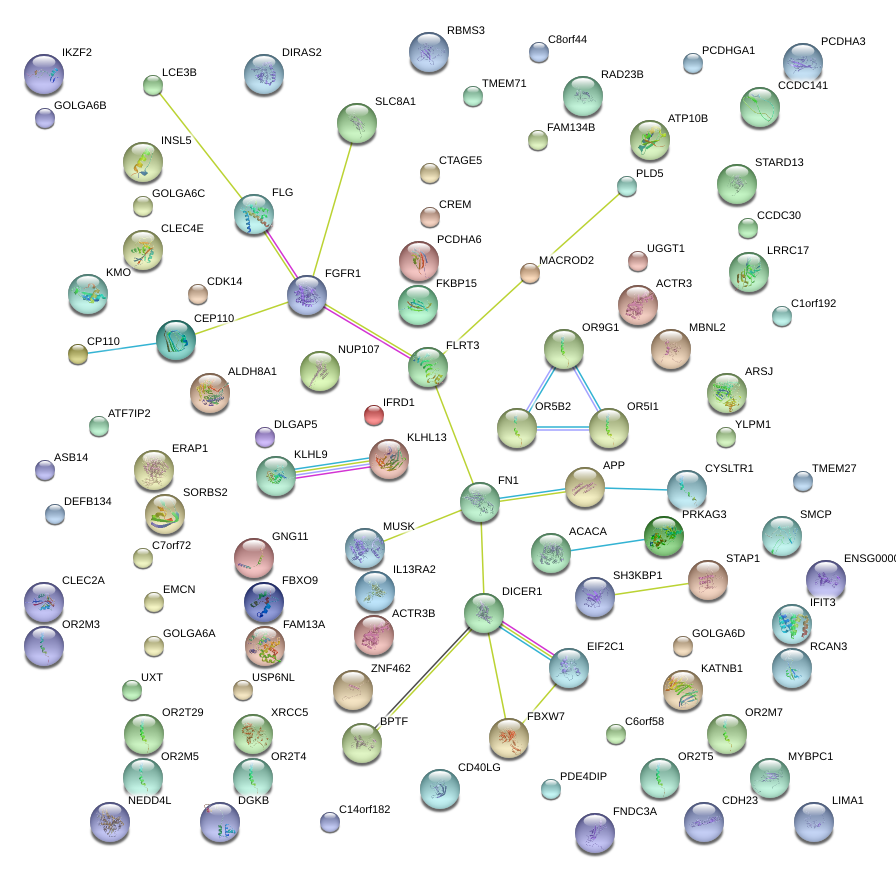
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr14_-_49543987
|
1.583
|
NM_001012706
|
C14orf182
|
chromosome 14 open reading frame 182
|
|
chr4_-_89963177
|
1.568
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr5_+_140690387
|
1.489
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr8_-_11891168
|
1.474
|
NM_001033019
|
DEFB134
|
defensin, beta 134
|
|
chr7_+_111879348
|
1.445
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr9_-_115077504
|
1.444
|
|
FKBP15
|
FK506 binding protein 15, 133kDa
|
|
chr5_-_16561936
|
1.437
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr6_+_53043729
|
1.436
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr4_-_153493133
|
1.431
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chrX_-_117003310
|
1.431
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr1_+_151117421
|
1.429
|
NM_030663
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr1_+_42773146
|
1.421
|
NM_001080850
|
CCDC30
|
coiled-coil domain containing 30
|
|
chrX_-_77469635
|
1.416
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr14_+_74353417
|
1.414
|
|
YLPM1
|
YLP motif containing 1
|
|
chr1_-_150564302
|
1.403
|
NM_002016
|
FLG
|
filaggrin
|
|
chr15_+_73337951
|
1.403
|
NM_001164404
|
GOLGA6A
GOLGA6C
|
golgin A6 family, member A
golgin A6 family, member C
|
|
chr1_+_24713396
|
1.398
|
|
RCAN3
|
RCAN family member 3
|
|
chr7_+_93388951
|
1.385
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr9_+_108665168
|
1.381
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr4_-_101658094
|
1.343
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr1_-_246554492
|
1.332
|
NM_001004691
|
OR2M7
|
olfactory receptor, family 2, subfamily M, member 7
|
|
chr20_-_14266200
|
1.328
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr1_-_159604287
|
1.316
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr1_-_246789396
|
1.304
|
NM_001004694
|
OR2T29
OR2T5
|
olfactory receptor, family 2, subfamily T, member 29
olfactory receptor, family 2, subfamily T, member 5
|
|
chr10_+_73225533
|
1.300
|
NM_001171933
NM_001171934
|
CDH23
|
cadherin-related 23
|
|
chr11_-_55460451
|
1.299
|
NM_006637
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1
|
|
chrX_-_19598912
|
1.280
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_-_115120251
|
1.275
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr3_+_73193499
|
1.274
|
NM_018029
|
EBLN2
|
endogenous Borna-like N element-2
|
|
chr1_-_93172119
|
1.241
|
|
|
|
|
chr4_-_186814852
|
1.231
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_+_216682365
|
1.227
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chr10_+_35524835
|
1.218
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr2_-_179623030
|
1.203
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr2_+_128644510
|
1.191
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr10_-_11614279
|
1.181
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr9_+_112470871
|
1.178
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr18_+_54013575
|
1.178
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr7_+_102340579
|
1.169
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr17_-_32730804
|
1.168
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr12_-_8584658
|
1.156
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr12_+_100512877
|
1.145
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr1_-_67039518
|
1.145
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr8_+_67751007
|
1.141
|
NM_019607
|
C8orf44
|
chromosome 8 open reading frame 44
|
|
chr5_-_160044844
|
1.140
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_-_48963505
|
1.130
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_+_36160375
|
1.125
|
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr12_-_9976173
|
1.124
|
NM_001130711
|
CLEC2A
|
C-type lectin domain family 2, member A
|
|
chr2_-_219404755
|
1.115
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr5_-_96175560
|
1.102
|
NM_001198541
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr1_-_143786993
|
1.102
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_-_57947358
|
1.099
|
NM_001005566
|
OR5B2
|
olfactory receptor, family 5, subfamily B, member 2
|
|
chr6_-_135312899
|
1.099
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr21_-_26294332
|
1.097
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr14_-_54720101
|
1.088
|
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr2_+_114390975
|
1.086
|
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast)
|
|
chr2_-_40593074
|
1.082
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chrX_+_135558001
|
1.076
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr13_+_96592051
|
1.067
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_-_32678142
|
1.065
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_+_56224436
|
1.059
|
NM_001005213
NM_001013358
|
OR9G1
OR9G9
|
olfactory receptor, family 9, subfamily G, member 1
olfactory receptor, family 9, subfamily G, member 9
|
|
chr16_+_56327168
|
1.057
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr9_+_122890394
|
1.051
|
NM_007018
|
CEP110
|
centrosomal protein 110kDa
|
|
chr20_+_15125503
|
1.050
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_-_215953818
|
1.036
|
|
FN1
|
fibronectin 1
|
|
chr16_+_10430225
|
1.035
|
NM_024997
|
ATF7IP2
|
activating transcription factor 7 interacting protein 2
|
|
chr8_-_133841988
|
1.031
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr7_+_90177082
|
1.028
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_+_91654395
|
1.026
|
NM_001004330
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7
|
|
chr1_+_169421011
|
1.024
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr4_+_124537334
|
1.024
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr3_+_1109619
|
1.020
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr2_-_175170674
|
1.012
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr11_-_58102169
|
1.012
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr14_+_55148488
|
1.010
|
|
KTN1
|
kinectin 1 (kinesin receptor)
|
|
chr4_-_38482805
|
1.010
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr8_+_107529327
|
1.010
|
NM_001198532
|
OXR1
|
oxidation resistance 1
|
|
chr14_+_98247702
|
1.009
|
NM_182560
|
C14orf177
|
chromosome 14 open reading frame 177
|
|
chr11_+_27019084
|
1.005
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr9_-_115077644
|
0.999
|
NM_139286
|
CDC26
|
cell division cycle 26 homolog (S. cerevisiae)
|
|
chr5_-_137876416
|
0.997
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr12_+_102505180
|
0.995
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr17_-_36779399
|
0.986
|
NM_002279
|
KRT33B
|
keratin 33B
|
|
chr10_-_21475493
|
0.982
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr15_-_75259714
|
0.980
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr2_+_202379442
|
0.979
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr7_+_93862137
|
0.978
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr14_+_38720085
|
0.975
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr18_+_30589937
|
0.974
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_+_88024152
|
0.973
|
|
ZNF292
|
zinc finger protein 292
|
|
chr9_-_21177529
|
0.972
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr2_+_38858830
|
0.964
|
NM_024775
|
GEMIN6
|
gem (nuclear organelle) associated protein 6
|
|
chr4_-_100431093
|
0.964
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr12_+_53699995
|
0.957
|
NM_021191
|
NEUROD4
|
neurogenic differentiation 4
|
|
chr1_+_86785346
|
0.951
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr1_+_63861474
|
0.950
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr11_+_118259684
|
0.949
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr7_+_90176647
|
0.946
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr11_-_102156531
|
0.946
|
NM_002425
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr11_+_20005704
|
0.944
|
|
NAV2
|
neuron navigator 2
|
|
chr5_+_140532385
|
0.943
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr11_+_55776251
|
0.940
|
NM_001004747
|
OR5T3
|
olfactory receptor, family 5, subfamily T, member 3
|
|
chr3_+_50119261
|
0.939
|
|
RBM5
|
RNA binding motif protein 5
|
|
chr9_+_676640
|
0.932
|
|
|
|
|
chrX_+_11687655
|
0.927
|
NM_006800
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr12_-_132042920
|
0.926
|
NM_183238
NM_001164715
|
ZNF605
|
zinc finger protein 605
|
|
chr9_-_28709302
|
0.920
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr4_-_120462705
|
0.920
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr1_+_47261826
|
0.919
|
NM_178033
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1
|
|
chr4_+_38740246
|
0.919
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr7_+_141124710
|
0.913
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chr3_-_52688768
|
0.910
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr2_-_101134146
|
0.908
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr18_+_3443771
|
0.904
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_+_19807168
|
0.898
|
|
|
|
|
chr5_-_92942680
|
0.897
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr12_-_48902692
|
0.897
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr6_-_117096594
|
0.894
|
NM_145062
|
ZUFSP
|
zinc finger with UFM1-specific peptidase domain
|
|
chr20_-_17589122
|
0.890
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr2_-_179380301
|
0.885
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr2_+_191009265
|
0.883
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr3_-_194579207
|
0.881
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr10_-_102269580
|
0.880
|
NM_015490
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chr1_+_161305558
|
0.876
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_228386800
|
0.873
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr7_-_64088848
|
0.872
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr17_-_30783631
|
0.872
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr17_-_36192311
|
0.871
|
NM_181537
|
KRT27
|
keratin 27
|
|
chr5_-_121440704
|
0.869
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr6_+_106653429
|
0.869
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_+_34993234
|
0.867
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr4_+_141484046
|
0.864
|
NM_001153446
NM_001153635
NM_001153585
NM_001153690
|
SCOC
|
short coiled-coil protein
|
|
chr17_-_36436979
|
0.859
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr1_-_21310462
|
0.855
|
NM_003760
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr11_+_124984559
|
0.851
|
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr10_+_90336489
|
0.851
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr2_+_207115060
|
0.843
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr17_-_38150551
|
0.842
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr12_+_34066482
|
0.841
|
NM_032834
|
ALG10
|
asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe)
|
|
chr5_+_148711419
|
0.839
|
|
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli)
|
|
chr1_-_56817802
|
0.839
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr4_+_71126376
|
0.829
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr2_-_172458970
|
0.829
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr8_-_82606104
|
0.824
|
NM_001105281
|
FABP12
|
fatty acid binding protein 12
|
|
chr1_-_144779601
|
0.824
|
NM_001101663
|
NBPF11
NBPF24
|
neuroblastoma breakpoint family, member 11
neuroblastoma breakpoint family, member 24
|
|
chr5_+_92944680
|
0.822
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr17_+_57811645
|
0.819
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr22_-_41672990
|
0.818
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr14_+_88130482
|
0.818
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr3_+_186563529
|
0.813
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr11_-_95715976
|
0.810
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr13_+_107753764
|
0.805
|
|
|
|
|
chr12_-_10497195
|
0.802
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr12_-_51153788
|
0.801
|
NM_173086
|
KRT6C
|
keratin 6C
|
|
chr17_-_3142625
|
0.796
|
NM_002550
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1
|
|
chr9_-_72673753
|
0.791
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr4_+_6726699
|
0.788
|
|
LOC93622
|
hypothetical LOC93622
|
|
chr18_-_19496590
|
0.788
|
NM_173505
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr18_+_27281729
|
0.787
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr7_+_156677777
|
0.787
|
|
|
|
|
chr4_+_159662349
|
0.786
|
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr12_+_27568311
|
0.785
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr1_+_78888064
|
0.779
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr9_-_130830255
|
0.778
|
NM_020145
|
SH3GLB2
|
SH3-domain GRB2-like endophilin B2
|
|
chr5_+_167651063
|
0.777
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr13_-_33148899
|
0.777
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_138132006
|
0.777
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr2_-_73373956
|
0.774
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr10_+_95838801
|
0.772
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr10_+_5976354
|
0.771
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr5_-_135729062
|
0.768
|
NM_001167576
NM_001167577
NM_020389
|
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7
|
|
chr2_+_113911737
|
0.767
|
NM_172003
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr1_+_51340487
|
0.767
|
NM_001136508
|
C1orf185
|
chromosome 1 open reading frame 185
|
|
chr11_-_55757236
|
0.765
|
NM_001004746
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2
|
|
chr3_+_176059763
|
0.762
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr7_-_36730525
|
0.761
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr13_-_40735619
|
0.759
|
NM_004294
|
MTRF1
|
mitochondrial translational release factor 1
|