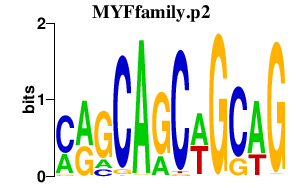
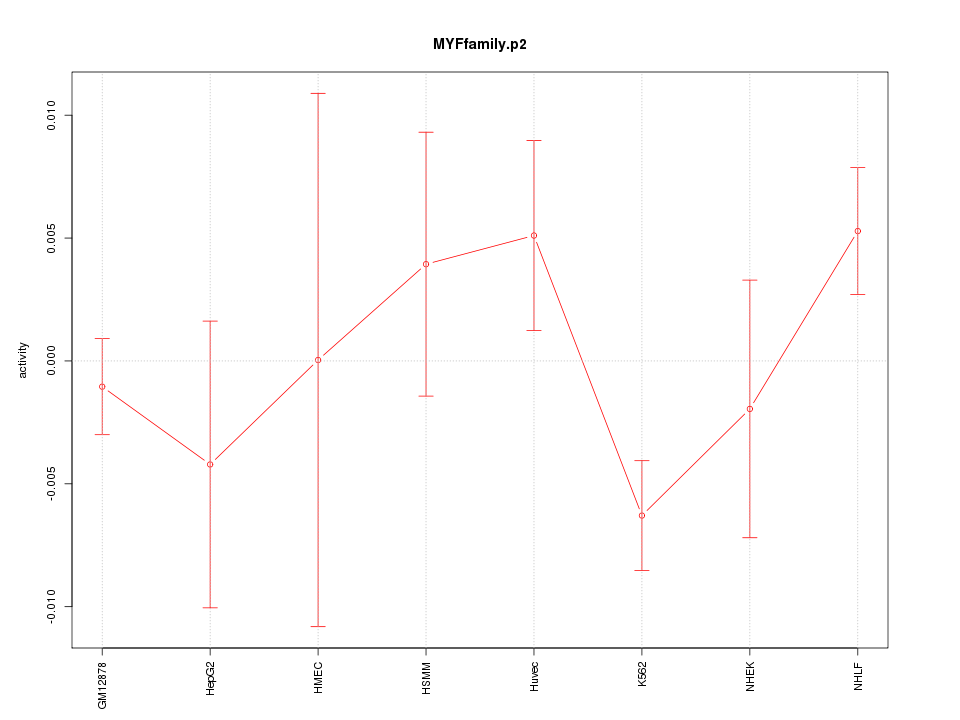
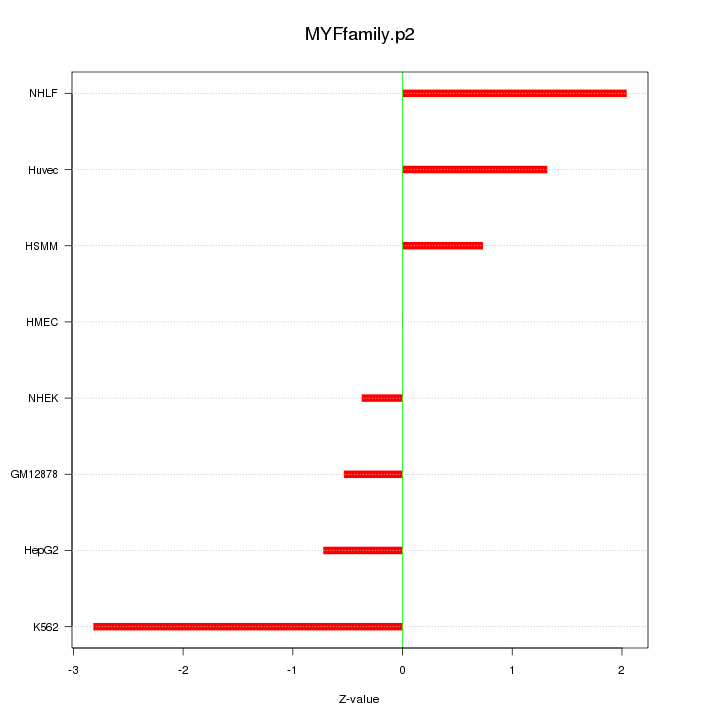
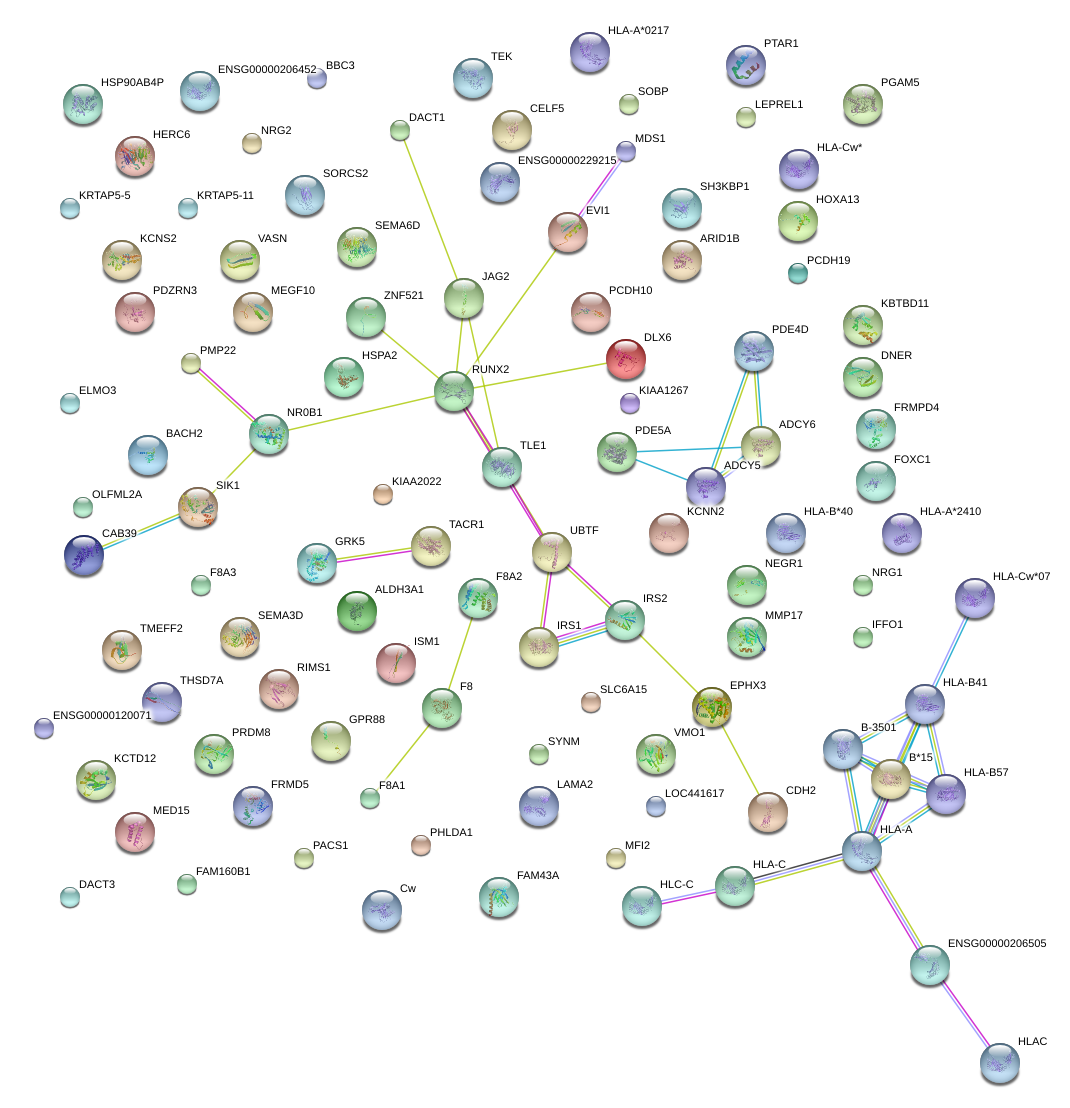
|
chr5_-_139402909
|
5.664
|
|
NRG2
|
neuregulin 2
|
|
chr5_-_139402989
|
3.639
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr8_+_31616809
|
2.647
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr3_-_191321601
|
2.568
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr1_-_72521079
|
2.514
|
|
|
|
|
chr6_+_45498354
|
2.305
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr6_+_45498262
|
2.116
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr1_-_72520735
|
2.059
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr17_-_15104768
|
2.022
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chrX_-_19815586
|
1.860
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chrX_-_19815526
|
1.851
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr18_-_24010944
|
1.827
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr3_+_195889845
|
1.740
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr10_+_120957084
|
1.734
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr7_-_84654106
|
1.702
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr7_-_11838260
|
1.617
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr4_-_120769275
|
1.562
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr6_+_107917965
|
1.559
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr6_+_129245978
|
1.548
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr6_+_157141565
|
1.539
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr6_+_1556543
|
1.527
|
|
FOXC1
|
forkhead box C1
|
|
chr19_-_52426055
|
1.488
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr12_-_6535481
|
1.485
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr3_-_170864111
|
1.464
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chrX_-_99551926
|
1.463
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr6_-_91063181
|
1.442
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr3_-_191321367
|
1.439
|
|
LEPREL1
|
leprecan-like 1
|
|
chr15_+_45796802
|
1.421
|
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr17_-_19589363
|
1.411
|
NM_001135168
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1
|
|
chr3_-_124650081
|
1.395
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr2_-_192767844
|
1.382
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_-_19815370
|
1.377
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chrX_-_154341451
|
1.368
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr2_-_39040981
|
1.345
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr10_+_116571485
|
1.343
|
NM_001135051
NM_020940
|
FAM160B1
|
family with sequence similarity 160, member B1
|
|
chr12_-_83830710
|
1.333
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chrX_-_30237403
|
1.327
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr4_+_134292630
|
1.324
|
|
PCDH10
|
protocadherin 10
|
|
chr5_-_59225377
|
1.314
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_100777044
|
1.301
|
|
GPR88
|
G protein-coupled receptor 88
|
|
chr2_-_230287442
|
1.282
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr12_-_74710741
|
1.270
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr21_-_43671355
|
1.266
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr8_+_1937711
|
1.264
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr12_+_6915698
|
1.261
|
|
|
|
|
chr12_+_130878870
|
1.258
|
NM_016155
|
MMP17
|
matrix metallopeptidase 17 (membrane-inserted)
|
|
chr16_+_65790514
|
1.247
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr9_-_83494198
|
1.243
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr20_+_13150417
|
1.240
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr17_-_39652449
|
1.235
|
NM_001076684
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr15_+_97462675
|
1.233
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr16_+_4370879
|
1.220
|
|
VASN
|
vasorin
|
|
chr9_+_126579203
|
1.219
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr5_+_113725914
|
1.217
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr12_-_83830700
|
1.184
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr4_+_89518898
|
1.177
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr7_+_96473225
|
1.176
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr12_-_83830670
|
1.172
|
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr14_+_58174489
|
1.165
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr4_+_7245218
|
1.160
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr11_+_65594361
|
1.154
|
NM_018026
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr18_-_21185968
|
1.150
|
|
ZNF521
|
zinc finger protein 521
|
|
chrX_-_74062005
|
1.137
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr4_-_120769590
|
1.136
|
|
|
|
|
chr3_-_73756668
|
1.109
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr22_+_19248733
|
1.106
|
|
MED15
|
mediator complex subunit 15
|
|
chr5_+_126654354
|
1.095
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr7_-_27206249
|
1.066
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr17_-_4636248
|
1.053
|
NM_001144939
NM_001144940
NM_001144941
NM_182566
|
VMO1
|
vitelline membrane outer layer 1 homolog (chicken)
|
|
chr14_-_104705753
|
1.050
|
|
JAG2
|
jagged 2
|
|
chrX_+_12066505
|
1.048
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr4_+_89519004
|
1.043
|
|
HERC6
|
hect domain and RLD 6
|
|
chr17_-_39652391
|
1.032
|
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr14_+_64076916
|
1.021
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr2_-_227372574
|
1.007
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr2_-_75280121
|
1.007
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr19_+_3175700
|
0.994
|
NM_021938
NM_001172673
|
CELF5
|
CUGBP, Elav-like family member 5
|
|
chr8_+_99508425
|
0.991
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr17_-_41625921
|
0.986
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr3_+_198214173
|
0.983
|
|
LOC100507057
MFI2
|
hypothetical LOC100507057
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr13_-_76358370
|
0.977
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr15_-_42274529
|
0.961
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr17_-_41626032
|
0.959
|
NM_015443
|
KIAA1267
|
KIAA1267
|
|
chr13_-_109236897
|
0.954
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr4_+_81325447
|
0.952
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr11_-_70971556
|
0.949
|
NM_001005405
|
KRTAP5-11
|
keratin associated protein 5-11
|
|
chr4_+_159311353
|
0.948
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr11_+_74629597
|
0.946
|
NM_001195528
|
LOC441617
|
hypothetical LOC441617
|
|
chr9_+_27099138
|
0.908
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr14_-_60817621
|
0.901
|
|
|
|
|
chr19_-_15204194
|
0.894
|
NM_024794
|
EPHX3
|
epoxide hydrolase 3
|
|
chr2_+_231285868
|
0.892
|
|
CAB39
|
calcium binding protein 39
|
|
chr6_+_72653126
|
0.886
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_-_31347867
|
0.885
|
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chr10_+_52504239
|
0.883
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr14_-_60817663
|
0.882
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr1_-_199704773
|
0.875
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr4_+_134293171
|
0.873
|
|
PCDH10
|
protocadherin 10
|
|
chr3_+_141136896
|
0.872
|
|
CLSTN2
|
calsyntenin 2
|
|
chr4_-_57671307
|
0.872
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr18_-_21186107
|
0.869
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr7_+_98084531
|
0.862
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr18_-_24011349
|
0.859
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr4_+_89519134
|
0.856
|
|
HERC6
|
hect domain and RLD 6
|
|
chr4_-_57671273
|
0.848
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr12_-_88269884
|
0.843
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr21_-_31853160
|
0.840
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr14_+_64949447
|
0.837
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr22_-_49311722
|
0.832
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr6_-_31347805
|
0.828
|
NM_002117
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chr1_-_32176562
|
0.826
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr19_+_54884252
|
0.816
|
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr12_-_93568336
|
0.815
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr16_-_2710552
|
0.814
|
NM_031948
|
PRSS27
|
protease, serine 27
|
|
chr9_-_130980302
|
0.810
|
|
IER5L
|
immediate early response 5-like
|
|
chr1_+_144273662
|
0.810
|
|
ANKRD35
|
ankyrin repeat domain 35
|
|
chr21_+_41461962
|
0.800
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr6_+_124166767
|
0.798
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chrX_+_153767803
|
0.795
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr13_+_34948885
|
0.788
|
|
NBEA
|
neurobeachin
|
|
chr15_-_53822468
|
0.784
|
NM_173814
|
PRTG
|
protogenin
|
|
chr14_-_74148422
|
0.783
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr21_-_27139143
|
0.781
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr17_+_23394062
|
0.778
|
|
NLK
|
nemo-like kinase
|
|
chr17_+_1906262
|
0.777
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr3_+_141136691
|
0.767
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr9_-_25668230
|
0.765
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr3_-_191522886
|
0.764
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr8_-_23768242
|
0.762
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr7_-_100068612
|
0.760
|
|
TFR2
|
transferrin receptor 2
|
|
chr7_-_102070473
|
0.757
|
NM_001114403
|
UPK3BL
POLR2J2
|
uroplakin 3B-like
polymerase (RNA) II (DNA directed) polypeptide J2
|
|
chr1_-_32176475
|
0.754
|
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr1_-_29321599
|
0.752
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr10_+_90629880
|
0.752
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr20_-_43953036
|
0.750
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr14_-_60817709
|
0.749
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr10_-_90957028
|
0.748
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr5_-_147142457
|
0.745
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_-_87122958
|
0.737
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr19_+_3175734
|
0.735
|
|
CELF5
|
CUGBP, Elav-like family member 5
|
|
chr7_-_138316393
|
0.732
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr2_+_233417348
|
0.727
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr4_-_86106567
|
0.720
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr4_-_57671112
|
0.718
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr15_+_97009194
|
0.718
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr14_-_60817946
|
0.717
|
|
|
|
|
chr3_-_87122936
|
0.716
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr8_+_60194328
|
0.715
|
|
|
|
|
chr4_-_25473528
|
0.712
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr8_-_52884456
|
0.703
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr12_+_64504835
|
0.703
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr4_-_75183700
|
0.702
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr11_+_64635870
|
0.696
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr9_+_136673534
|
0.692
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr4_-_75183860
|
0.690
|
NM_002089
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr2_+_36436873
|
0.685
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr5_-_158459006
|
0.681
|
|
EBF1
|
early B-cell factor 1
|
|
chr4_-_186814852
|
0.679
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_19155360
|
0.676
|
NM_001136265
|
IFFO2
|
intermediate filament family orphan 2
|
|
chr5_-_147142444
|
0.675
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr12_+_6807831
|
0.675
|
NM_014262
|
LEPREL2
|
leprecan-like 2
|
|
chr22_+_19248810
|
0.674
|
|
MED15
|
mediator complex subunit 15
|
|
chr14_-_104706159
|
0.668
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr9_+_130223046
|
0.667
|
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chrX_-_46503415
|
0.666
|
NM_032591
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr17_-_4640335
|
0.665
|
|
LOC284014
|
hypothetical protein LOC284014
|
|
chr1_-_150044365
|
0.663
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr8_-_72918697
|
0.663
|
|
MSC
|
musculin
|
|
chr16_-_66571863
|
0.661
|
NM_001129758
NM_022357
|
DPEP3
|
dipeptidase 3
|
|
chr2_-_197883703
|
0.657
|
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr1_+_110555433
|
0.654
|
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr9_+_86474457
|
0.654
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr5_+_129268352
|
0.653
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr9_+_22437064
|
0.650
|
|
|
|
|
chr1_+_219119623
|
0.645
|
|
HLX
|
H2.0-like homeobox
|
|
chr15_-_42274760
|
0.645
|
|
FRMD5
|
FERM domain containing 5
|
|
chr1_+_153232685
|
0.642
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr10_-_118022965
|
0.640
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr9_+_86474393
|
0.635
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr2_-_227371698
|
0.631
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr15_+_89229275
|
0.631
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr9_+_86474500
|
0.629
|
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr14_-_50631794
|
0.628
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr3_-_45242072
|
0.623
|
|
TMEM158
|
transmembrane protein 158 (gene/pseudogene)
|
|
chr1_-_33613729
|
0.620
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr3_+_160964574
|
0.611
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_+_142252890
|
0.610
|
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr6_-_32299757
|
0.609
|
NM_004557
|
NOTCH4
|
notch 4
|
|
chr4_-_186815116
|
0.603
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_+_108665168
|
0.601
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr17_-_7434113
|
0.599
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr8_+_22078622
|
0.598
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr22_-_49311439
|
0.595
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr21_+_41461597
|
0.593
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr1_+_67163165
|
0.588
|
NM_001077700
NM_001077702
NM_001077703
NM_001146110
NM_001146111
NM_001146112
NM_001146113
NM_020948
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr9_+_86474445
|
0.587
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr12_+_120055013
|
0.586
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr7_-_122313646
|
0.585
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr19_-_13478037
|
0.585
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr1_+_211190631
|
0.578
|
|
VASH2
|
vasohibin 2
|