|
chr14_-_105900687
|
4.260
|
|
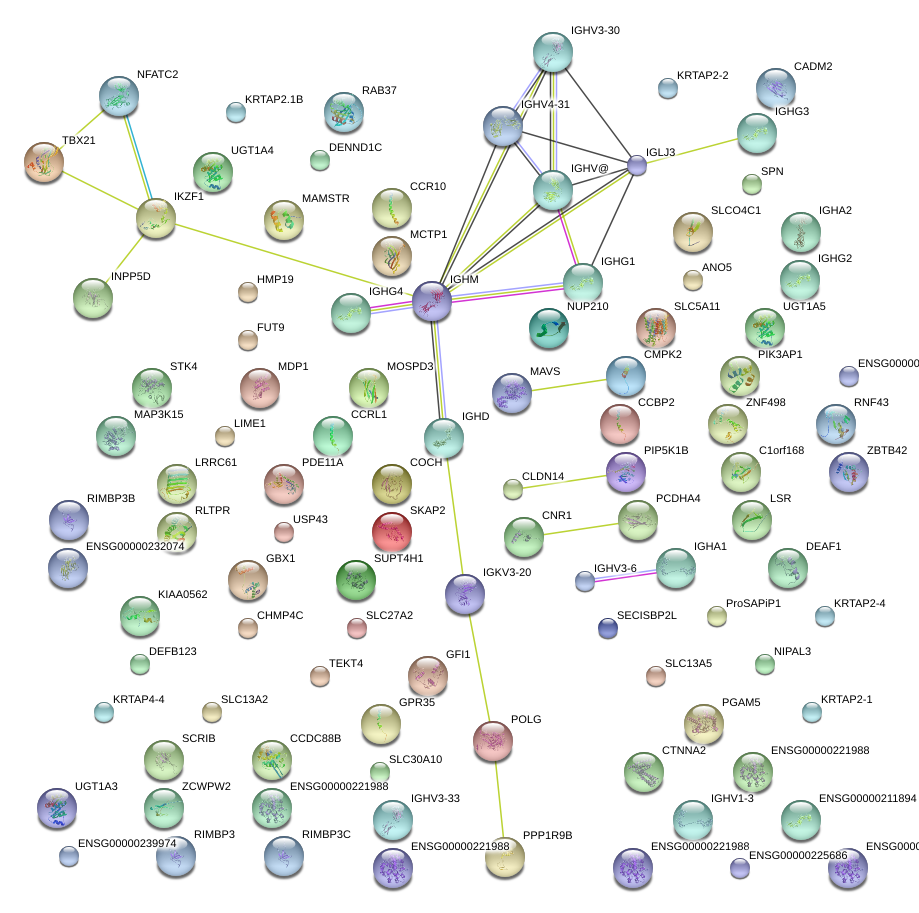
IGHA1
IGHD
IGHG3
IGHM
SKAP2
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
src kinase associated phosphoprotein 2
|
|
chr22_-_20235372
|
3.406
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr14_-_106202262
|
3.037
|
|
IGLJ3
IGHA1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
|
|
chr10_-_98470164
|
2.904
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr22_+_20068039
|
2.885
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr14_-_106084085
|
2.696
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_94646033
|
2.647
|
NM_024717
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr20_-_3097070
|
2.506
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr6_+_96570565
|
2.493
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr14_-_23754977
|
2.299
|
NM_138476
|
MDP1
|
magnesium-dependent phosphatase 1
|
|
chr14_-_105851580
|
2.271
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_-_13436802
|
2.214
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr14_-_105862080
|
2.182
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr17_+_70244553
|
2.175
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr5_+_173405343
|
2.136
|
|
HMP19
|
HMP19 protein
|
|
chr11_+_22171150
|
2.125
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr17_+_23824784
|
2.103
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr6_+_32230172
|
2.076
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr1_-_92724181
|
2.071
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr14_-_105589467
|
2.066
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr11_+_63864263
|
2.004
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr19_+_40431122
|
1.996
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr20_+_29492071
|
1.953
|
NM_153324
|
DEFB123
|
defensin, beta 123
|
|
chr19_-_53914768
|
1.910
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_-_218168137
|
1.890
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr14_-_106282128
|
1.868
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr19_+_40431359
|
1.852
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr9_+_70509925
|
1.850
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr20_-_49592654
|
1.780
|
NM_012340
NM_173091
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr14_-_106119742
|
1.760
|
|
IGLJ3
IGHG1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_+_43165608
|
1.744
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr21_-_36774257
|
1.731
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr20_+_3775421
|
1.697
|
NM_020746
|
MAVS
|
mitochondrial antiviral signaling protein
|
|
chr14_-_105785639
|
1.693
|
|
|
|
|
chr20_-_49592444
|
1.684
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr14_-_106289751
|
1.580
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr17_+_9489578
|
1.577
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chrX_-_19443299
|
1.566
|
NM_001001671
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15
|
|
chr8_+_82807218
|
1.498
|
NM_152284
|
CHMP4C
|
chromatin modifying protein 4C
|
|
chr14_-_105796268
|
1.471
|
|
ZCWPW2
IGHV3-23
IGKV3-20
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
|
|
chr16_+_29582480
|
1.464
|
|
SPN
|
sialophorin
|
|
chr2_+_234302511
|
1.449
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr14_-_106154332
|
1.446
|
|
LOC100126583
IGHG1
|
hypothetical LOC100126583
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_-_36456949
|
1.438
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr16_+_24765052
|
1.410
|
NM_052944
|
SLC5A11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11
|
|
chr5_-_101659864
|
1.410
|
NM_180991
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1
|
|
chr14_+_30413393
|
1.402
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr5_+_173405298
|
1.396
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr19_+_40431618
|
1.394
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_+_233633279
|
1.357
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr6_-_88911708
|
1.350
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr2_+_233633348
|
1.345
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr7_+_100047877
|
1.345
|
NM_001040098
NM_001040099
NM_023948
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr2_-_178645718
|
1.341
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr17_+_70244945
|
1.340
|
NM_001006638
NM_001163990
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr3_+_85090721
|
1.328
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr16_+_66236559
|
1.286
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr2_+_94900904
|
1.284
|
NM_144705
|
TEKT4
|
tektin 4
|
|
chr15_+_48261649
|
1.279
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr14_-_106352471
|
1.279
|
|
|
|
|
chr1_-_3763620
|
1.251
|
NM_014704
|
KIAA0562
|
KIAA0562
|
|
chr19_-_6432765
|
1.251
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr2_+_241193492
|
1.248
|
NM_001195381
NM_001195382
|
GPR35
|
G protein-coupled receptor 35
|
|
chr7_-_150495799
|
1.247
|
NM_001098834
|
GBX1
|
gastrulation brain homeobox 1
|
|
chr12_+_131797455
|
1.212
|
NM_001170543
NM_001170544
NM_138575
|
PGAM5
|
phosphoglycerate mutase family member 5
|
|
chr2_-_6923215
|
1.202
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr20_+_43028533
|
1.199
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr20_+_61839931
|
1.195
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr17_-_53847937
|
1.188
|
|
RNF43
|
ring finger protein 43
|
|
chr14_+_104337923
|
1.181
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr7_+_50314801
|
1.177
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr14_-_105762742
|
1.174
|
|
|
|
|
chr2_+_79593567
|
1.165
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr17_-_36570508
|
1.164
|
NM_032524
|
KRTAP4-4
|
keratin associated protein 4-4
|
|
chr1_+_24614831
|
1.162
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr1_-_57057956
|
1.160
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chr20_+_43028590
|
1.159
|
|
STK4
|
serine/threonine kinase 4
|
|
chr15_-_47125788
|
1.158
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr7_+_149651202
|
1.155
|
NM_001142928
NM_023942
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr17_-_53784494
|
1.155
|
NM_003168
|
SUPT4H1
|
suppressor of Ty 4 homolog 1 (S. cerevisiae)
|
|
chr8_-_144969529
|
1.150
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr7_+_99052489
|
1.145
|
NM_145115
|
ZNF498
|
zinc finger protein 498
|
|
chr17_-_38087366
|
1.139
|
NM_016602
|
CCR10
|
chemokine (C-C motif) receptor 10
|
|
chr17_-_4410565
|
1.127
|
NM_001122890
NM_153338
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr2_+_233881047
|
1.127
|
NM_000541
|
SAG
|
S-antigen; retina and pineal gland (arrestin)
|
|
chr9_-_129869211
|
1.126
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr1_-_24614173
|
1.117
|
NM_001199012
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr14_+_23675233
|
1.115
|
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr22_+_38213174
|
1.113
|
NM_001098270
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr19_-_15451203
|
1.103
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr17_-_34015631
|
1.100
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr20_+_17628592
|
1.091
|
NM_001014977
NM_178477
|
BANF2
|
barrier to autointegration factor 2
|
|
chr19_-_48700824
|
1.089
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr14_+_23675206
|
1.078
|
NM_006263
NM_176783
|
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha)
|
|
chr7_+_149651481
|
1.075
|
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr20_+_56848189
|
1.072
|
NM_016592
|
GNAS
|
GNAS complex locus
|
|
chr6_+_87703742
|
1.071
|
NM_000865
|
HTR1E
|
5-hydroxytryptamine (serotonin) receptor 1E
|
|
chr1_-_40903897
|
1.049
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr1_-_16217025
|
1.045
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr1_-_40903700
|
1.044
|
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr14_-_105644300
|
1.044
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr18_-_45975163
|
1.035
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr6_+_35373632
|
1.035
|
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr1_+_109827975
|
1.032
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr19_-_8670054
|
1.031
|
NM_178525
|
ACTL9
|
actin-like 9
|
|
chr1_-_3763480
|
1.029
|
|
KIAA0562
|
KIAA0562
|
|
chr1_-_229181219
|
1.025
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr2_-_108972100
|
1.022
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr11_+_63829616
|
1.019
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chrX_-_150893752
|
1.012
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr20_+_56848165
|
1.011
|
|
GNAS
|
GNAS complex locus
|
|
chr1_-_229181240
|
1.010
|
NM_001122835
NM_024525
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr14_-_23981493
|
1.006
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr1_-_229181229
|
1.005
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr7_-_86526880
|
1.002
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr6_+_35373572
|
1.001
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr17_-_37221443
|
0.995
|
|
|
|
|
chr13_+_99431417
|
0.979
|
|
|
|
|
chr19_+_5774817
|
0.979
|
NM_004558
|
NRTN
|
neurturin
|
|
chr2_-_108972048
|
0.978
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr20_+_56848127
|
0.976
|
|
GNAS
|
GNAS complex locus
|
|
chr12_+_124044187
|
0.967
|
|
BRI3BP
|
BRI3 binding protein
|
|
chr3_-_50335113
|
0.965
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr5_+_140997171
|
0.965
|
|
RELL2
|
RELT-like 2
|
|
chr20_+_60744268
|
0.959
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr6_+_32229771
|
0.951
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr12_+_124044104
|
0.948
|
NM_080626
|
BRI3BP
|
BRI3 binding protein
|
|
chr7_+_129913271
|
0.946
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr17_-_36533903
|
0.945
|
NM_031854
|
KRTAP4-12
|
keratin associated protein 4-12
|
|
chr17_-_38385891
|
0.942
|
NM_001136042
NM_001142653
NM_001142654
|
AARSD1
|
alanyl-tRNA synthetase domain containing 1
|
|
chr20_+_36988469
|
0.942
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr3_+_39826032
|
0.941
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr16_+_66236512
|
0.934
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr7_-_149651594
|
0.932
|
NM_001164458
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast)
|
|
chr3_+_50167424
|
0.932
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr11_+_817584
|
0.930
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr2_+_79593679
|
0.927
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr15_-_19974402
|
0.926
|
|
|
|
|
chr7_-_100009205
|
0.921
|
NM_001168682
|
SAP25
|
sin3A-binding protein 25
|
|
chr8_-_139995417
|
0.920
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr16_-_88295595
|
0.919
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr17_+_38386107
|
0.915
|
NM_173079
|
RUNDC1
|
RUN domain containing 1
|
|
chr17_+_38386113
|
0.913
|
|
RUNDC1
|
RUN domain containing 1
|
|
chr20_+_36663865
|
0.913
|
NM_001164431
|
ARHGAP40
|
Rho GTPase activating protein 40
|
|
chr17_+_16259636
|
0.906
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr21_-_36836767
|
0.899
|
NM_001146078
|
CLDN14
|
claudin 14
|
|
chr14_-_105845570
|
0.897
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr16_+_23754997
|
0.889
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_-_226661089
|
0.887
|
NM_145214
|
TRIM11
|
tripartite motif containing 11
|
|
chr6_-_5952631
|
0.883
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr9_+_139292094
|
0.880
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr14_-_105712630
|
0.880
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_-_6557375
|
0.870
|
NM_001143838
NM_177550
|
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5
|
|
chr4_-_40326603
|
0.869
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr4_-_159311959
|
0.860
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_-_77647505
|
0.856
|
|
FASN
|
fatty acid synthase
|
|
chr1_+_17571277
|
0.848
|
NM_207421
|
PADI6
|
peptidyl arginine deiminase, type VI
|
|
chr20_+_36988368
|
0.846
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr2_+_219453487
|
0.845
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr3_-_69064364
|
0.843
|
NM_001005527
NM_182522
|
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4
|
|
chr16_-_30032330
|
0.838
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr2_+_171999082
|
0.836
|
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr1_+_15935395
|
0.836
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr2_-_70634129
|
0.828
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr1_-_229181203
|
0.825
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr10_+_80735881
|
0.824
|
|
|
|
|
chr20_+_36867708
|
0.823
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr9_+_134843918
|
0.821
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr1_+_18680010
|
0.821
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr6_-_33493792
|
0.818
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr14_+_23654307
|
0.817
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr14_-_22358752
|
0.813
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr12_+_56135353
|
0.809
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr10_+_94823636
|
0.808
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr1_+_27062215
|
0.808
|
NM_006142
|
SFN
|
stratifin
|
|
chr4_+_159311353
|
0.806
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr17_+_16259573
|
0.801
|
NM_016113
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr1_-_23729871
|
0.801
|
|
E2F2
|
E2F transcription factor 2
|
|
chr1_-_109651140
|
0.797
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr22_+_21742364
|
0.792
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr12_+_6819635
|
0.791
|
NM_002075
|
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3
|
|
chr5_+_142130475
|
0.785
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr11_+_77452550
|
0.783
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr7_+_119700924
|
0.782
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr3_+_141879555
|
0.774
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr20_-_51644187
|
0.773
|
|
ZNF217
|
zinc finger protein 217
|
|
chr1_-_2486313
|
0.773
|
NM_003820
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator)
|
|
chr19_+_14405216
|
0.772
|
|
PKN1
|
protein kinase N1
|
|
chr19_+_18069596
|
0.769
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr16_-_8870343
|
0.767
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr1_+_229181462
|
0.766
|
|
ARV1
|
ARV1 homolog (S. cerevisiae)
|
|
chr6_+_31791063
|
0.758
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr16_+_29582069
|
0.757
|
NM_003123
|
SPN
|
sialophorin
|
|
chr16_-_87244896
|
0.751
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr17_+_71033339
|
0.747
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr12_-_49878216
|
0.746
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr2_+_171999042
|
0.744
|
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr10_+_94823221
|
0.743
|
NM_057157
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr22_+_20880112
|
0.742
|
|
|
|
|
chr6_-_33493898
|
0.740
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|