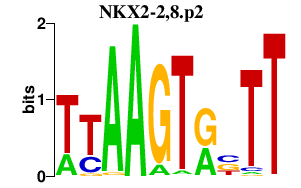
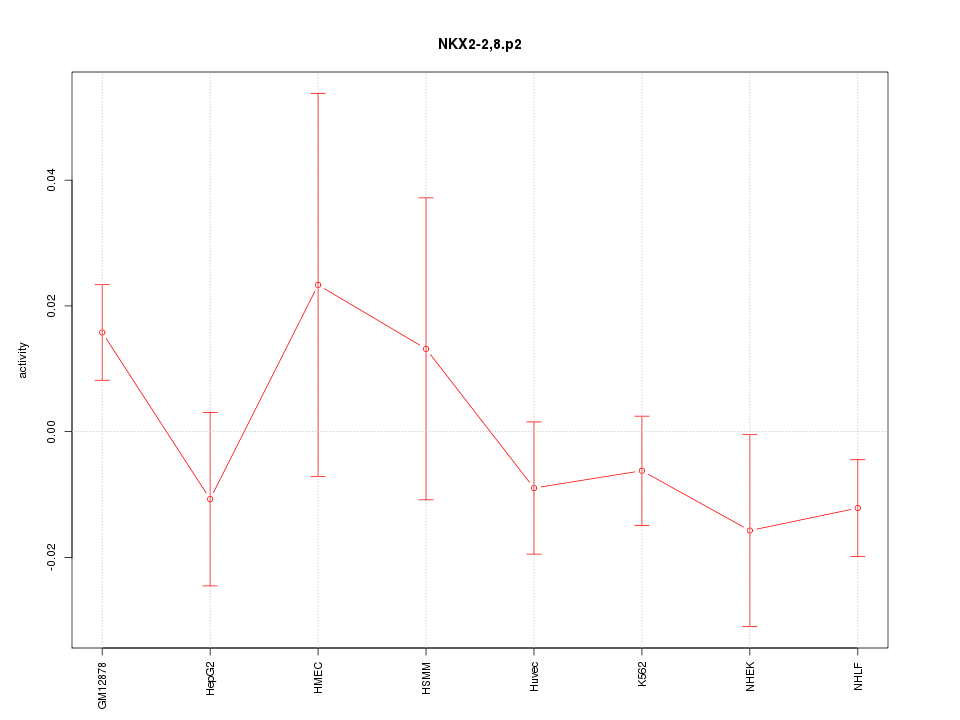
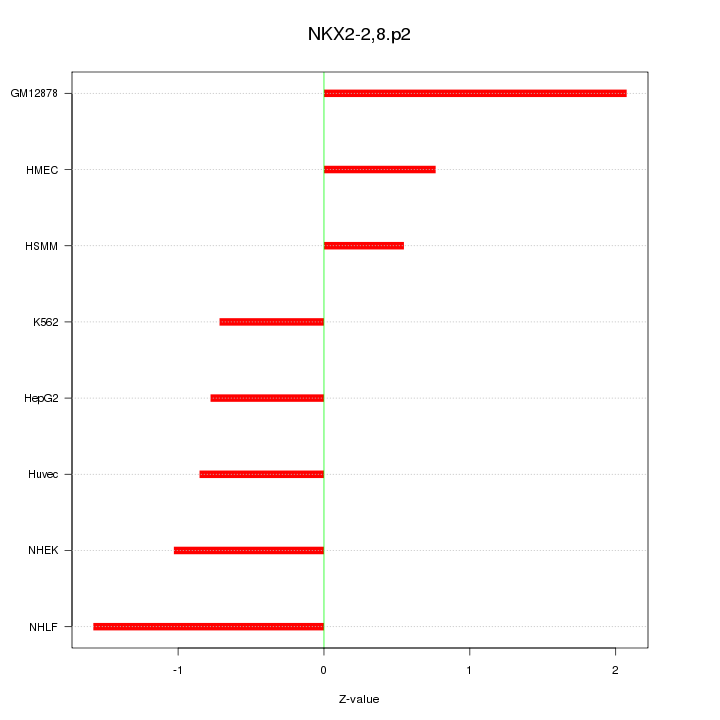
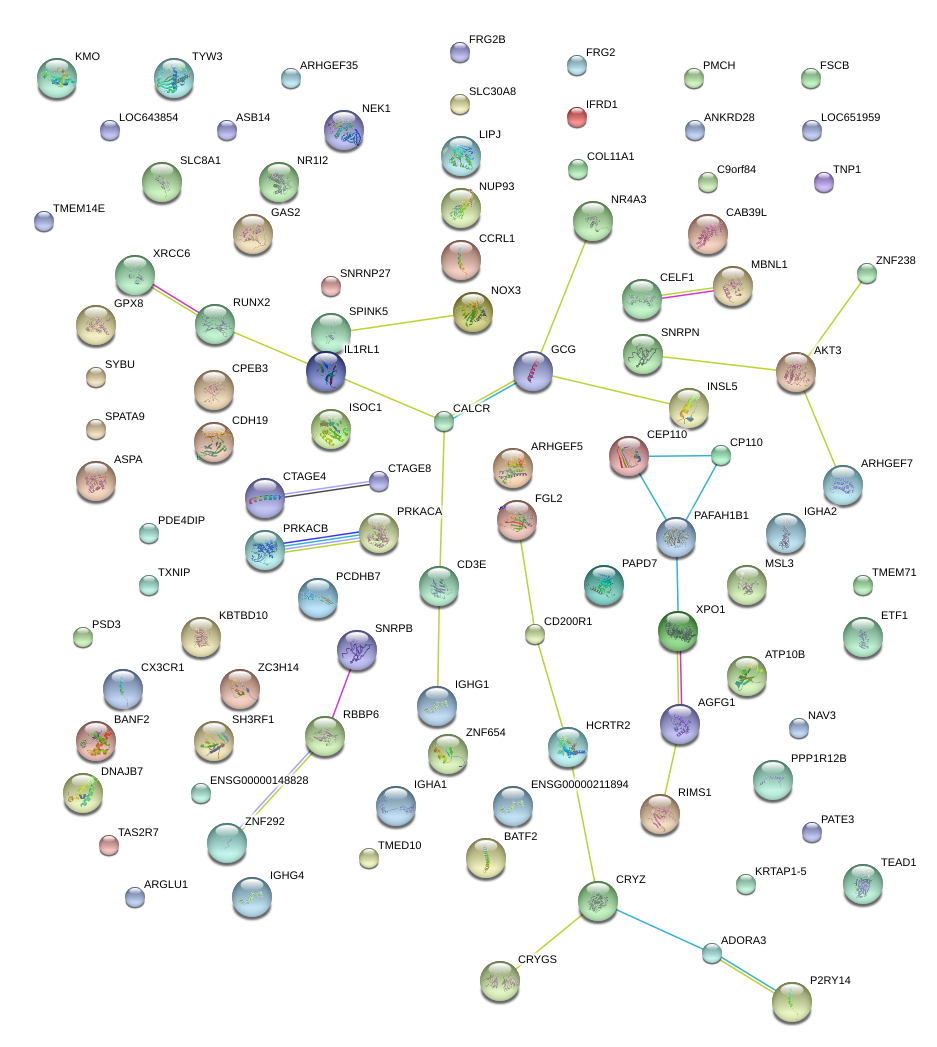
|
chr7_+_37689903
|
4.124
|
|
|
|
|
chr7_+_111879348
|
2.977
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr5_-_160044844
|
2.951
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr9_+_122923864
|
2.827
|
|
CEP110
|
centrosomal protein 110kDa
|
|
chr1_+_239762179
|
2.462
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr10_-_93992982
|
2.027
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr1_-_74971679
|
1.968
|
NM_001130042
NM_001130043
NM_001134759
NM_001889
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr20_+_17622319
|
1.857
|
NM_001159495
|
BANF2
|
barrier to autointegration factor 2
|
|
chr11_+_117680504
|
1.774
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr12_-_101115677
|
1.731
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr11_+_65027721
|
1.713
|
|
|
|
|
chr3_-_57301749
|
1.666
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr2_-_61566477
|
1.624
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chrX_+_11687655
|
1.550
|
NM_006800
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr3_+_153499883
|
1.522
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr4_-_170770111
|
1.507
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr3_-_15814678
|
1.506
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr16_+_24475129
|
1.486
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_-_143708467
|
1.480
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_103269356
|
1.461
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr9_-_113561613
|
1.437
|
NM_001080551
|
C9orf84
|
chromosome 9 open reading frame 84
|
|
chr6_-_155818728
|
1.392
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr5_-_137876416
|
1.374
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr17_+_3324096
|
1.369
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr1_+_200698493
|
1.343
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr1_+_74971423
|
1.324
|
NM_001162916
NM_138467
|
TYW3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae)
|
|
chr14_-_44046202
|
1.309
|
NM_032135
|
FSCB
|
fibrous sheath CABYR binding protein
|
|
chr7_-_76667045
|
1.308
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr3_+_133798770
|
1.304
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr3_-_152449591
|
1.264
|
NM_001081455
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr3_+_88270951
|
1.258
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr2_+_69974581
|
1.243
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr6_+_55147029
|
1.241
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr11_+_12917369
|
1.241
|
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr1_-_111908085
|
1.233
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr3_-_121065112
|
1.227
|
|
|
|
|
chr4_-_170314382
|
1.226
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr9_-_3213007
|
1.216
|
|
|
|
|
chr1_-_197173159
|
1.204
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr5_+_128468785
|
1.194
|
|
ISOC1
|
isochorismatase domain containing 1
|
|
chr1_-_67039518
|
1.180
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr2_-_162717159
|
1.152
|
NM_002054
|
GCG
|
glucagon
|
|
chr3_+_120982020
|
1.129
|
NM_003889
NM_033013
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr12_-_10846435
|
1.124
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr10_+_90336489
|
1.119
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr5_+_147423727
|
1.115
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr15_-_75364387
|
1.093
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr2_-_40593074
|
1.092
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr15_+_22778233
|
1.024
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr1_+_242281175
|
1.022
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr5_+_6799100
|
1.021
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr2_+_102320148
|
1.016
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr6_+_73133152
|
1.006
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_-_153541468
|
1.003
|
NM_001123228
|
TMEM14E
|
transmembrane protein 14E
|
|
chr1_+_15862887
|
0.993
|
|
|
|
|
chr8_+_118216517
|
0.982
|
NM_001172814
NM_173851
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8
|
|
chr11_-_47467094
|
0.975
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr10_-_37931864
|
0.966
|
NM_001190489
|
MTRNR2L7
|
MT-RNR2-like 7
|
|
chr5_+_54491702
|
0.962
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr15_+_22832198
|
0.962
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr17_-_1195096
|
0.949
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr2_-_217433018
|
0.948
|
NM_003284
|
TNP1
|
transition protein 1 (during histone to protamine replacement)
|
|
chr8_-_133841988
|
0.938
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr16_+_55373241
|
0.938
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr6_+_88021644
|
0.931
|
|
ZNF292
|
zinc finger protein 292
|
|
chr14_-_106184988
|
0.929
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_+_125163215
|
0.926
|
NM_001129883
|
PATE3
|
prostate and testis expressed 3
|
|
chr6_+_45404031
|
0.917
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr22_+_40372957
|
0.916
|
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr1_+_84402962
|
0.915
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr5_-_95044415
|
0.913
|
NM_031952
|
SPATA9
|
spermatogenesis associated 9
|
|
chr1_+_169747788
|
0.907
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr13_-_48873735
|
0.906
|
NM_030925
|
CAB39L
|
calcium binding protein 39-like
|
|
chr8_-_110689396
|
0.897
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr14_+_88130482
|
0.876
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr14_-_31026901
|
0.873
|
NM_001197184
|
GPR33
|
G protein-coupled receptor 33 (gene/pseudogene)
|
|
chr7_-_143597313
|
0.859
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr14_-_74669864
|
0.857
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr8_-_18710575
|
0.856
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_+_63510481
|
0.849
|
|
|
|
|
chr18_-_62422195
|
0.835
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr3_-_187744808
|
0.833
|
NM_017541
|
CRYGS
|
crystallin, gamma S
|
|
chr1_+_144149794
|
0.828
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr22_-_39587944
|
0.821
|
NM_145174
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7
|
|
chr2_+_170074457
|
0.818
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr1_-_74971276
|
0.815
|
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chr2_+_228064531
|
0.812
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr17_-_36436979
|
0.812
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr10_-_135290288
|
0.808
|
NM_001080998
|
FRG2B
FRG2
|
FSHD region gene 2 family, member B
FSHD region gene 2
|
|
chr7_-_93027057
|
0.801
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chr12_+_76749199
|
0.792
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr13_-_106018445
|
0.778
|
NM_018011
|
ARGLU1
|
arginine and glutamate rich 1
|
|
chr11_+_22646229
|
0.774
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr13_+_110653407
|
0.773
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr9_+_101628829
|
0.771
|
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr5_+_140532385
|
0.767
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr1_-_242073094
|
0.767
|
NM_005465
NM_181690
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr21_+_24722913
|
0.756
|
|
FLJ42200
|
FLJ42200 protein
|
|
chr14_-_24148658
|
0.750
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr5_-_58688428
|
0.747
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_10850845
|
0.746
|
NM_023918
|
TAS2R8
|
taste receptor, type 2, member 8
|
|
chr12_+_32578513
|
0.737
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_+_75796170
|
0.730
|
NM_001124759
|
FRG2C
FLJ20518
|
FSHD region gene 2 family, member C
FSHD region gene 2 family, member C pseudogene
|
|
chr1_+_242281199
|
0.727
|
|
ZNF238
|
zinc finger protein 238
|
|
chr18_-_3835295
|
0.727
|
NM_001003809
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr14_-_81069865
|
0.716
|
|
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr3_-_152541129
|
0.714
|
NM_176876
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr12_+_54106304
|
0.706
|
NM_001005183
|
OR6C76
|
olfactory receptor, family 6, subfamily C, member 76
|
|
chr3_-_102194938
|
0.705
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr7_+_113853790
|
0.697
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr10_-_98326435
|
0.689
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr9_+_102380156
|
0.689
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr12_+_10054492
|
0.681
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chr9_-_21472311
|
0.681
|
NM_176891
|
IFNE
|
interferon, epsilon
|
|
chr18_-_65766068
|
0.679
|
|
CD226
|
CD226 molecule
|
|
chr3_-_69183926
|
0.675
|
NM_007114
|
TMF1
|
TATA element modulatory factor 1
|
|
chr1_+_144374513
|
0.671
|
|
RNF115
|
ring finger protein 115
|
|
chr12_+_63090008
|
0.670
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr5_-_169340321
|
0.667
|
NM_001129891
|
FAM196B
|
family with sequence similarity 196, member B
|
|
chr17_+_55098280
|
0.666
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr3_+_122769467
|
0.664
|
NM_001012659
|
ARGFX
|
arginine-fifty homeobox
|
|
chr2_-_225086564
|
0.663
|
|
CUL3
|
cullin 3
|
|
chr8_+_7742811
|
0.657
|
NM_058203
NM_001081552
|
SPAG11B
SPAG11A
|
sperm associated antigen 11B
sperm associated antigen 11A
|
|
chr6_-_41362370
|
0.651
|
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr3_-_122862447
|
0.651
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr10_-_98315090
|
0.650
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_20374273
|
0.648
|
NM_001105572
|
PLA2G2C
|
phospholipase A2, group IIC
|
|
chr4_+_88147107
|
0.642
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr1_+_154573728
|
0.639
|
NM_144627
|
C1orf182
|
chromosome 1 open reading frame 182
|
|
chr8_-_81155564
|
0.638
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr3_-_113701073
|
0.636
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr5_-_147996769
|
0.631
|
NM_001040169
NM_001040172
NM_001040173
NM_199453
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr14_-_19871267
|
0.628
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr14_+_66901237
|
0.625
|
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa
|
|
chr1_-_41398126
|
0.614
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr10_+_63478974
|
0.614
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr9_-_74735669
|
0.613
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr1_+_213245507
|
0.612
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr3_-_56673113
|
0.610
|
NM_015224
|
C3orf63
|
chromosome 3 open reading frame 63
|
|
chr2_-_187422141
|
0.608
|
NM_182521
|
ZSWIM2
|
zinc finger, SWIM-type containing 2
|
|
chr3_-_122862391
|
0.605
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr12_-_110410730
|
0.603
|
|
ATXN2
|
ataxin 2
|
|
chr6_-_132073849
|
0.597
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chrX_-_44287032
|
0.597
|
NM_173794
|
FUNDC1
|
FUN14 domain containing 1
|
|
chr1_+_103840900
|
0.596
|
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3
|
|
chr20_+_20296744
|
0.595
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr7_+_93373755
|
0.594
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr11_-_22838547
|
0.593
|
NM_001195637
|
LOC100500938
|
hypothetical LOC100500938
|
|
chr6_-_123999640
|
0.591
|
NM_006073
|
TRDN
|
triadin
|
|
chr3_-_33675484
|
0.591
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr4_+_69996780
|
0.584
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr12_+_100394787
|
0.584
|
NM_152323
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related)
|
|
chr7_-_92693716
|
0.581
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr7_+_90177082
|
0.577
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr10_+_90511142
|
0.576
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr13_-_98428244
|
0.576
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr4_-_157006827
|
0.572
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr4_-_153493133
|
0.571
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr4_+_109763119
|
0.570
|
|
RPL34
|
ribosomal protein L34
|
|
chr11_+_123315459
|
0.570
|
NM_001001965
|
OR4D5
|
olfactory receptor, family 4, subfamily D, member 5
|
|
chr1_-_156816312
|
0.566
|
NM_001004477
|
OR10X1
|
olfactory receptor, family 10, subfamily X, member 1
|
|
chr1_-_21249974
|
0.565
|
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr1_-_240679380
|
0.562
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr3_+_51550635
|
0.561
|
NM_015106
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae)
|
|
chr6_-_88095714
|
0.561
|
NM_198568
|
GJB7
|
gap junction protein, beta 7, 25kDa
|
|
chr4_-_177427252
|
0.559
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr8_+_17874128
|
0.553
|
|
PCM1
|
pericentriolar material 1
|
|
chr12_-_15706340
|
0.552
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_-_33393696
|
0.548
|
NM_001145338
|
ZBTB22
|
zinc finger and BTB domain containing 22
|
|
chr11_-_102001259
|
0.547
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr17_-_26648375
|
0.544
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr1_-_115039614
|
0.543
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr9_+_109131814
|
0.542
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr15_-_40537002
|
0.538
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr2_-_216008689
|
0.535
|
|
FN1
|
fibronectin 1
|
|
chr3_+_153014550
|
0.532
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr10_+_57028755
|
0.532
|
NM_001190478
|
MTRNR2L5
|
MT-RNR2-like 5
|
|
chr3_+_1109619
|
0.530
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr6_-_11490487
|
0.529
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr4_-_38534802
|
0.524
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr2_+_151922350
|
0.519
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr10_+_98730977
|
0.517
|
NM_015652
|
C10orf12
|
chromosome 10 open reading frame 12
|
|
chr11_-_5657947
|
0.516
|
|
TRIM5
|
tripartite motif containing 5
|
|
chr17_-_19290745
|
0.516
|
|
|
|
|
chr21_-_26175068
|
0.515
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr1_-_214963279
|
0.515
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_-_27569269
|
0.510
|
NM_145698
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr4_-_152368492
|
0.508
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_-_184478920
|
0.508
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr1_-_161379606
|
0.506
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr20_+_56659693
|
0.505
|
NM_001001433
NM_001134772
NM_001134773
NM_003763
|
STX16
|
syntaxin 16
|
|
chr21_-_30796278
|
0.505
|
NM_181611
|
KRTAP19-5
|
keratin associated protein 19-5
|
|
chr15_-_97366313
|
0.505
|
NM_001102612
|
PGPEP1L
|
pyroglutamyl-peptidase I-like
|
|
chr9_+_114671635
|
0.504
|
|
SNX30
|
sorting nexin family member 30
|
|
chr17_-_64462976
|
0.503
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr11_+_125258718
|
0.502
|
NM_145014
|
HYLS1
|
hydrolethalus syndrome 1
|
|
chr12_-_10215942
|
0.500
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr17_-_35327318
|
0.500
|
NM_018530
|
GSDMB
|
gasdermin B
|
|
chr2_-_20375497
|
0.496
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr17_-_35975227
|
0.496
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|