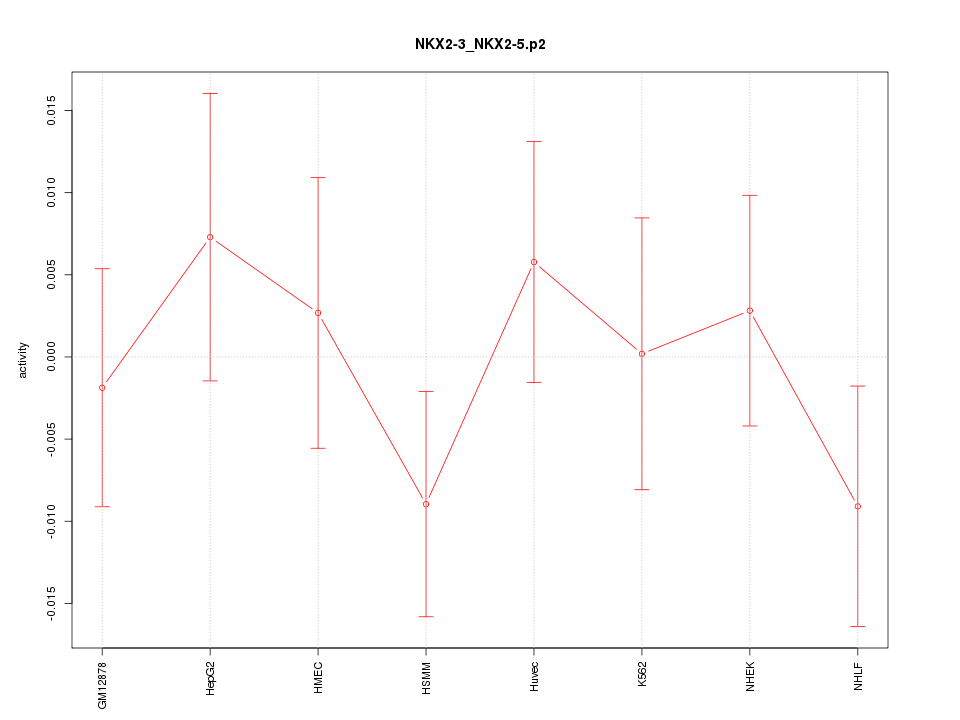
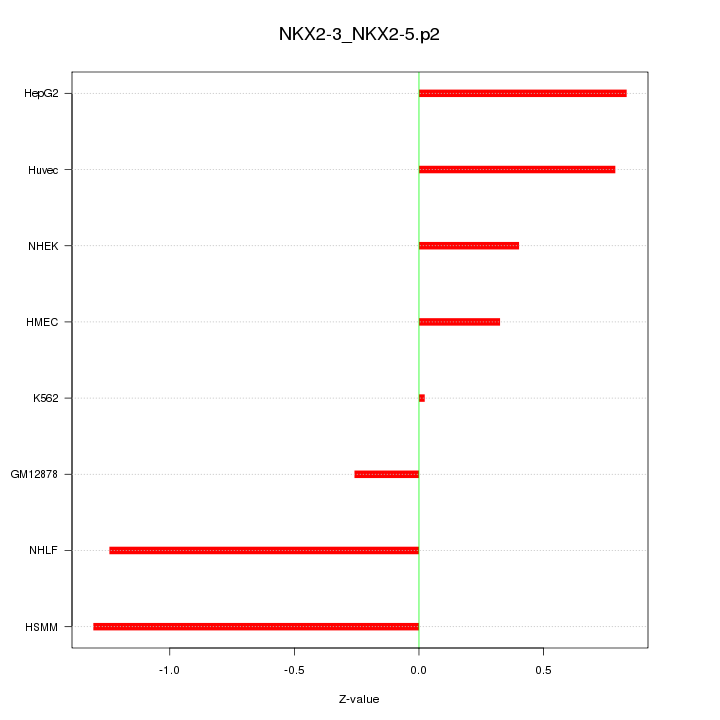
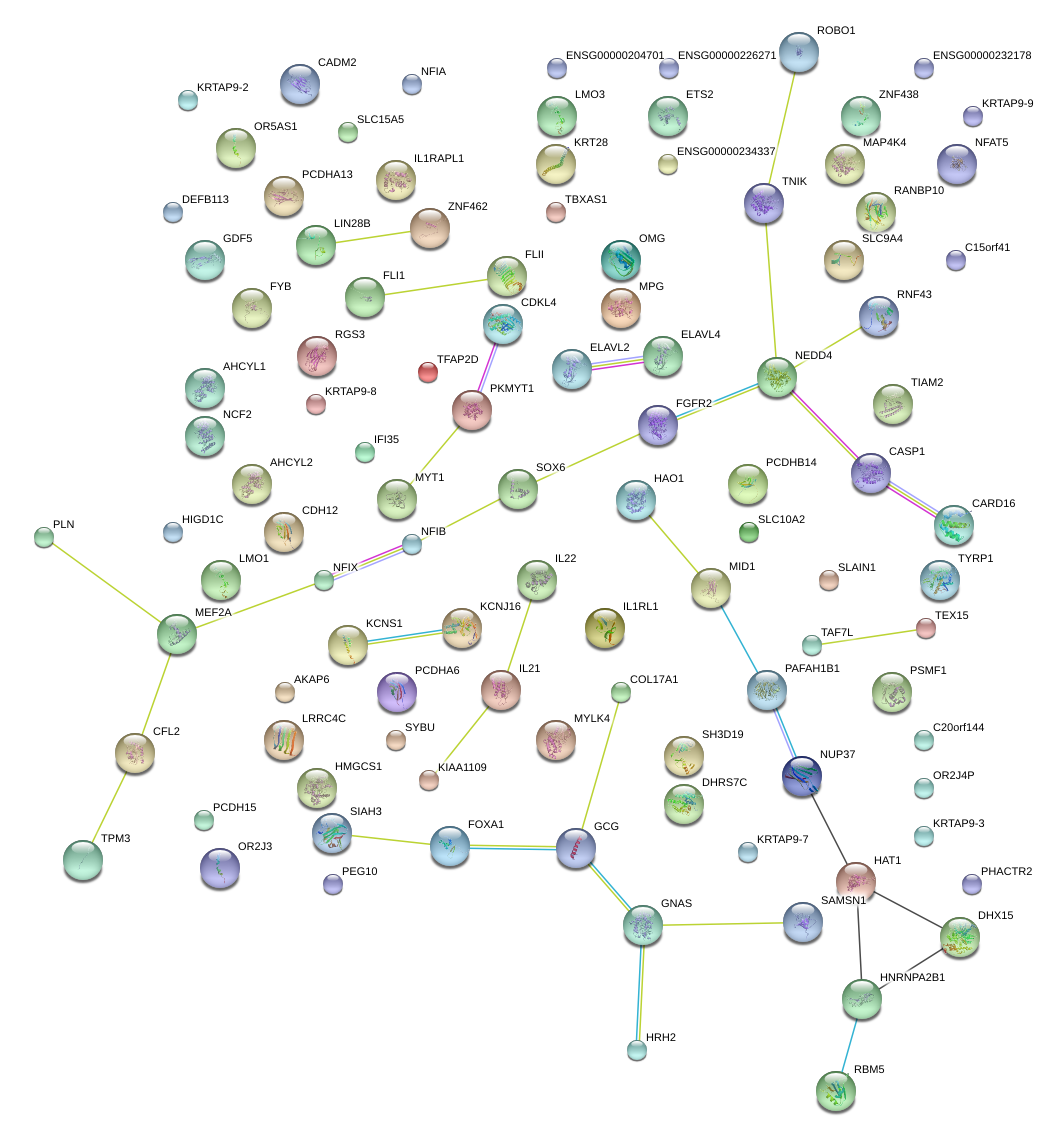
|
chr3_-_79899748
|
0.936
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr19_-_46504831
|
0.776
|
|
|
|
|
chr13_-_45323757
|
0.658
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr11_-_40272239
|
0.582
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr6_+_50789215
|
0.550
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr7_+_94135384
|
0.507
|
|
PEG10
|
paternally expressed 10
|
|
chr10_-_123346148
|
0.482
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr13_-_102517196
|
0.448
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr9_+_12683385
|
0.441
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr2_+_102456193
|
0.422
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr17_+_38412267
|
0.422
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chrX_-_10504852
|
0.410
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr13_+_77213295
|
0.399
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr6_+_29187565
|
0.396
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chr8_-_110724594
|
0.379
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr10_-_105835609
|
0.374
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr7_-_26202626
|
0.364
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr4_-_152368492
|
0.348
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_+_140227951
|
0.346
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr6_-_50045296
|
0.333
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chr4_-_123761615
|
0.326
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr7_-_26198673
|
0.325
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr14_-_34252695
|
0.323
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_+_7682355
|
0.322
|
|
|
|
|
chr2_+_37282516
|
0.307
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chr3_+_85858321
|
0.307
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr2_-_162717159
|
0.304
|
NM_002054
|
GCG
|
glucagon
|
|
chr10_-_105835525
|
0.302
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr5_+_175041069
|
0.298
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr17_-_1195096
|
0.282
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr20_-_7869068
|
0.278
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr20_-_33489383
|
0.276
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr5_-_22889487
|
0.275
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr11_+_128067598
|
0.275
|
NM_001167681
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr12_-_101036407
|
0.274
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr6_-_2696152
|
0.263
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr14_-_37134050
|
0.262
|
|
FOXA1
|
forkhead box A1
|
|
chr12_-_120725210
|
0.261
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr9_-_14076901
|
0.260
|
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_16652202
|
0.259
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr21_+_39117064
|
0.256
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr9_+_108726257
|
0.255
|
|
ZNF462
|
zinc finger protein 462
|
|
chr7_-_96471544
|
0.254
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr7_+_128802719
|
0.252
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_-_56230953
|
0.251
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr16_+_68238460
|
0.250
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr5_-_43326922
|
0.248
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr15_+_97990705
|
0.247
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr20_-_43163021
|
0.243
|
NM_002251
|
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1
|
|
chr15_-_53996597
|
0.239
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr17_-_9635325
|
0.235
|
NM_001105571
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr12_-_16321885
|
0.233
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr11_-_16380967
|
0.233
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_-_152431175
|
0.224
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr8_-_30826074
|
0.224
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr20_+_31713750
|
0.223
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr14_+_31868229
|
0.220
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr6_+_155579788
|
0.220
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr6_+_143971009
|
0.220
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr2_-_55053398
|
0.219
|
|
|
|
|
chr20_+_56919233
|
0.218
|
|
GNAS
|
GNAS complex locus
|
|
chr17_-_26648375
|
0.217
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr5_+_140583098
|
0.213
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr17_-_53849888
|
0.213
|
|
RNF43
|
ring finger protein 43
|
|
chr14_-_37133927
|
0.213
|
|
FOXA1
|
forkhead box A1
|
|
chr11_-_104411065
|
0.209
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr20_+_62266217
|
0.207
|
|
MYT1
|
myelin transcription factor 1
|
|
chr2_+_102320148
|
0.206
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr17_+_65582981
|
0.206
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr20_+_1041905
|
0.204
|
NM_178578
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr20_+_62266270
|
0.204
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr12_+_49634048
|
0.203
|
NM_001109619
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C
|
|
chr10_-_31328426
|
0.202
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr14_-_37134191
|
0.201
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr20_+_56904042
|
0.201
|
|
GNAS
|
GNAS complex locus
|
|
chr5_-_39255335
|
0.199
|
NM_001465
NM_199335
|
FYB
|
FYN binding protein
|
|
chr17_-_36209672
|
0.198
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr2_+_172530120
|
0.195
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr22_-_16127520
|
0.194
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr2_+_101874765
|
0.194
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_+_139175420
|
0.193
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr2_+_101874809
|
0.191
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr5_+_140215778
|
0.190
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr9_+_115303527
|
0.190
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chrX_+_28515479
|
0.190
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr14_+_22059212
|
0.189
|
|
|
|
|
chr15_+_34720097
|
0.186
|
NM_032499
|
C15orf41
|
chromosome 15 open reading frame 41
|
|
chr2_-_39310176
|
0.185
|
NM_001009565
|
CDKL4
|
cyclin-dependent kinase-like 4
|
|
chr1_-_181826633
|
0.185
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr4_+_123380569
|
0.183
|
|
KIAA1109
|
KIAA1109
|
|
chr6_+_118976134
|
0.183
|
NM_002667
|
PLN
|
phospholamban
|
|
chr11_+_55554470
|
0.183
|
NM_001001921
|
OR5AS1
|
olfactory receptor, family 5, subfamily AS, member 1
|
|
chr17_+_72065440
|
0.183
|
|
LOC100507246
|
hypothetical LOC100507246
|
|
chr21_-_14840506
|
0.181
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr1_+_50342168
|
0.178
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr6_+_105511615
|
0.178
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr3_+_50119261
|
0.176
|
|
RBM5
|
RNA binding motif protein 5
|
|
chr6_-_25679197
|
0.174
|
|
|
|
|
chr17_+_36642240
|
0.174
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr6_-_39398293
|
0.172
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr6_-_39180390
|
0.171
|
|
C6orf64
|
chromosome 6 open reading frame 64
|
|
chr1_+_63861474
|
0.171
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr4_+_68995761
|
0.171
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr17_-_36346240
|
0.170
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_-_142605739
|
0.170
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr17_+_55269974
|
0.170
|
|
|
|
|
chr12_-_10853940
|
0.170
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr20_+_30160952
|
0.169
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr5_+_140166842
|
0.169
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr17_-_39191658
|
0.167
|
NM_025237
|
SOST
|
sclerostin
|
|
chr6_-_66473790
|
0.165
|
NM_001142800
NM_001142801
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr5_-_160211623
|
0.165
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_+_12829807
|
0.163
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr10_+_124660206
|
0.161
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr14_-_105712630
|
0.160
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_-_92693716
|
0.160
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr2_-_175170674
|
0.160
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr14_+_30161285
|
0.159
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr12_-_4358968
|
0.159
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr11_-_57964221
|
0.158
|
NM_001004733
|
OR5B12
|
olfactory receptor, family 5, subfamily B, member 12
|
|
chr2_-_40593074
|
0.158
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr13_-_37070862
|
0.157
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr13_+_51933228
|
0.156
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr16_+_7322751
|
0.156
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr14_+_22059174
|
0.156
|
|
|
|
|
chr5_-_58331515
|
0.155
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_51200347
|
0.155
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr2_+_225973845
|
0.154
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr2_+_174907914
|
0.154
|
NM_001145250
|
SP9
|
Sp9 transcription factor homolog (mouse)
|
|
chr1_+_556968
|
0.154
|
|
|
|
|
chr2_-_162072492
|
0.153
|
|
AHCTF1P1
|
AT hook containing transcription factor 1 pseudogene 1
|
|
chr3_+_174598937
|
0.152
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr16_-_20246309
|
0.151
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr14_-_46882144
|
0.151
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr2_+_165804157
|
0.150
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr1_+_100089118
|
0.150
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr2_-_215953818
|
0.150
|
|
FN1
|
fibronectin 1
|
|
chr14_-_106289751
|
0.149
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr20_+_19985289
|
0.148
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr5_+_140703784
|
0.148
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr19_+_50100843
|
0.147
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr20_-_49747397
|
0.146
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr20_-_55368284
|
0.145
|
NM_001190472
|
MTRNR2L3
|
MT-RNR2-like 3
|
|
chr9_-_103185621
|
0.144
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr16_-_20323533
|
0.144
|
NM_174924
|
PDILT
|
protein disulfide isomerase-like, testis expressed
|
|
chr11_-_57173823
|
0.143
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_+_2528272
|
0.143
|
|
CNTN4
|
contactin 4
|
|
chr12_-_6355165
|
0.141
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr3_-_74652952
|
0.140
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr21_+_30734467
|
0.140
|
NM_181623
|
KRTAP15-1
|
keratin associated protein 15-1
|
|
chr10_+_102722275
|
0.140
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr3_-_121883647
|
0.140
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr14_+_38772870
|
0.140
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr2_-_207291364
|
0.139
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr8_+_104900591
|
0.139
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr10_+_115302767
|
0.139
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr4_+_75393053
|
0.138
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr8_-_20084892
|
0.138
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr10_-_98326435
|
0.137
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr12_+_15590552
|
0.137
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr5_-_168204430
|
0.137
|
|
|
|
|
chr20_+_5935897
|
0.137
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr2_+_234266250
|
0.136
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr10_-_21475493
|
0.135
|
NM_001010896
NM_001177483
|
C10orf113
|
chromosome 10 open reading frame 113
|
|
chr6_+_31731667
|
0.135
|
|
APOM
|
apolipoprotein M
|
|
chr17_-_64649608
|
0.135
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr6_+_31731647
|
0.135
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr20_-_29441946
|
0.135
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr2_+_137464931
|
0.133
|
NM_001080427
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr1_-_32157845
|
0.132
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr20_-_13919256
|
0.131
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr3_-_52269467
|
0.130
|
|
WDR82
|
WD repeat domain 82
|
|
chr3_-_115960807
|
0.130
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_-_36991189
|
0.130
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr6_-_50039776
|
0.130
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr2_-_20375497
|
0.130
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr12_-_23995109
|
0.129
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chrX_-_114158388
|
0.129
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr10_-_75071205
|
0.129
|
|
MYOZ1
|
myozenin 1
|
|
chr19_+_4442610
|
0.129
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr4_+_53986759
|
0.129
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr13_-_40454188
|
0.129
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr2_+_171280046
|
0.128
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr4_-_69218909
|
0.128
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr3_+_171560104
|
0.128
|
NM_001145097
|
SKIL
|
SKI-like oncogene
|
|
chr11_+_89032112
|
0.128
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr2_-_74460933
|
0.128
|
NM_001135040
NM_001190837
NM_004082
|
DCTN1
|
dynactin 1
|
|
chr4_-_69852090
|
0.127
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr8_+_9990471
|
0.127
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr9_+_135491305
|
0.127
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr8_-_122722674
|
0.126
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr1_+_157526127
|
0.126
|
NM_002001
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide
|
|
chr2_+_171280182
|
0.125
|
|
|
|
|
chr11_+_20001130
|
0.125
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr6_-_49712483
|
0.125
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr6_+_16251260
|
0.124
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr11_-_123319754
|
0.123
|
NM_001005187
|
OR6T1
|
olfactory receptor, family 6, subfamily T, member 1
|
|
chr12_-_27815475
|
0.123
|
NM_001146221
|
LOC100287284
|
MANSC domain-containing protein ENSP00000370673
|
|
chr10_+_118340460
|
0.123
|
NM_006229
|
PNLIPRP1
|
pancreatic lipase-related protein 1
|
|
chr12_-_6103945
|
0.122
|
NM_000552
|
VWF
|
von Willebrand factor
|