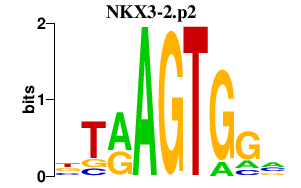
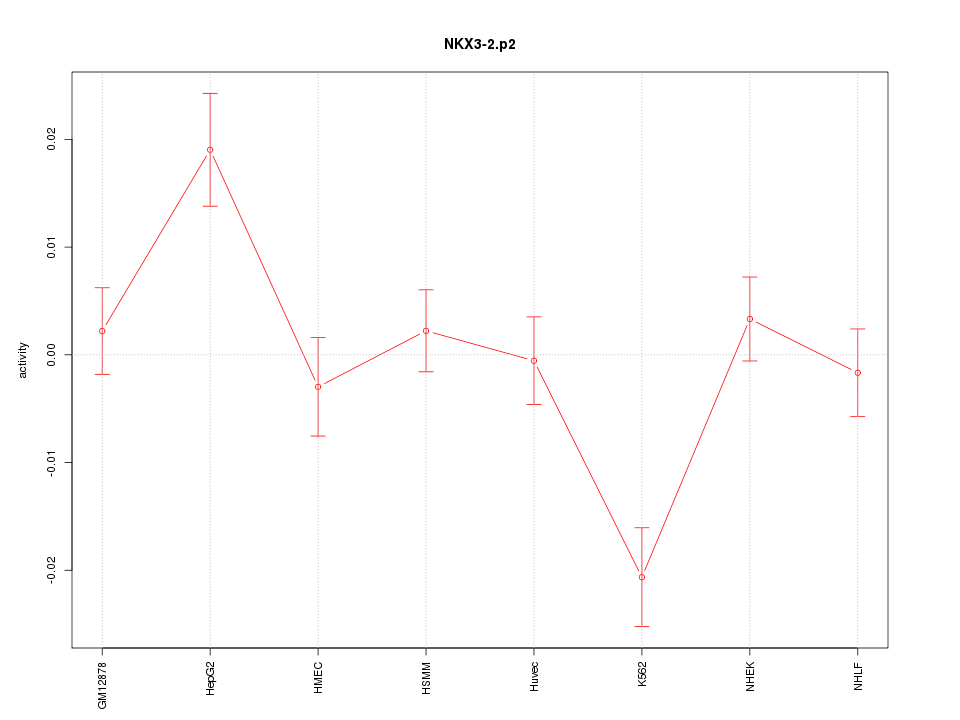
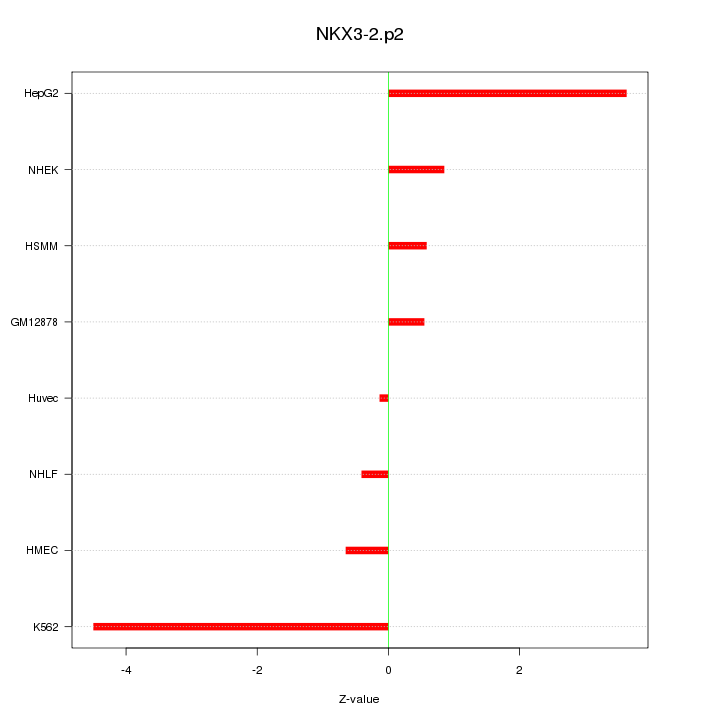
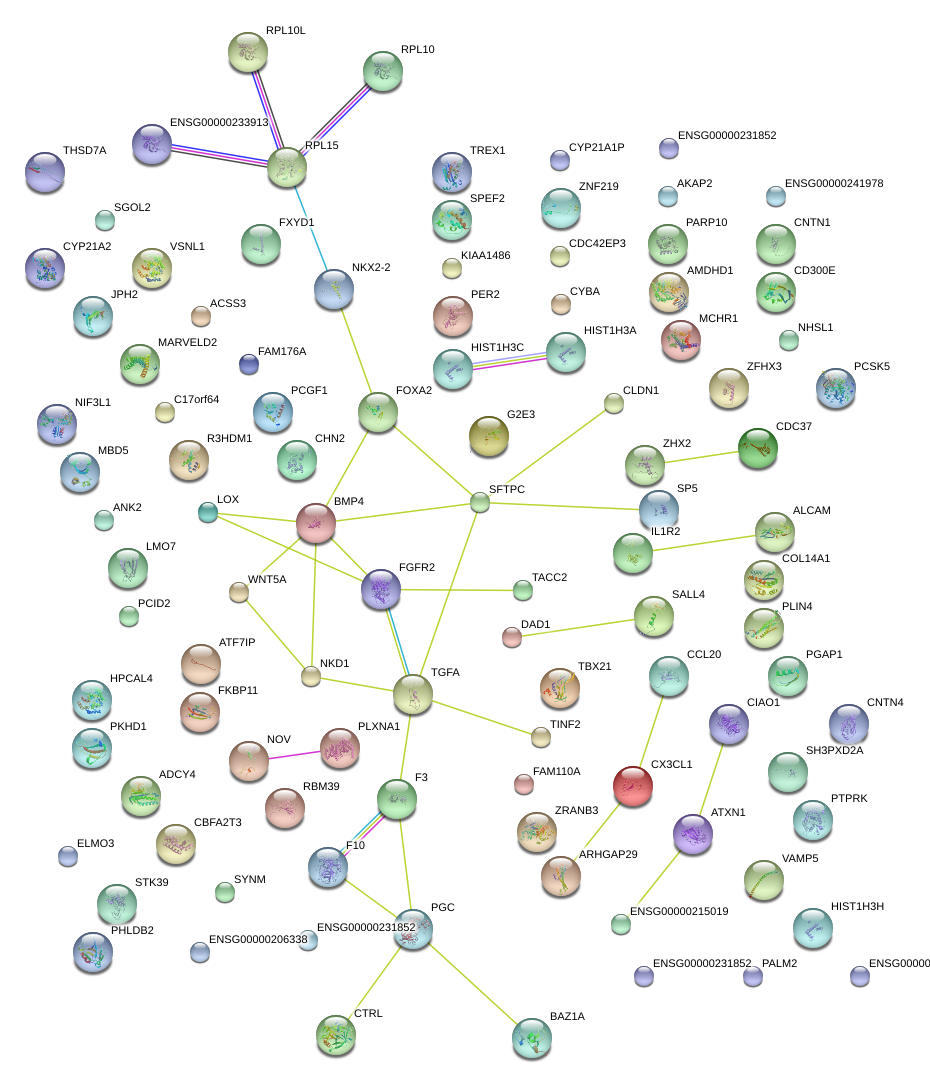
|
chr2_-_238861830
|
1.633
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr8_+_32905027
|
1.546
|
|
|
|
|
chr2_+_17585462
|
1.463
|
|
VSNL1
|
visinin-like 1
|
|
chr2_+_228386800
|
1.401
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr16_-_71650034
|
1.376
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr5_+_68746694
|
1.162
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr3_+_48482213
|
1.151
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr2_+_171280944
|
1.125
|
|
SP5
|
Sp5 transcription factor
|
|
chr16_+_55963883
|
1.111
|
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr14_+_61653827
|
1.102
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr16_+_55963899
|
1.026
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr14_-_20642214
|
1.010
|
|
|
|
|
chr2_+_17585287
|
0.982
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr2_-_136005238
|
0.972
|
NM_032143
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3
|
|
chr3_+_48482625
|
0.963
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr3_+_106568430
|
0.960
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr3_+_106568334
|
0.938
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr14_-_22840249
|
0.896
|
|
|
|
|
chr14_+_30098079
|
0.894
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr3_+_106568243
|
0.886
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr8_+_123863075
|
0.866
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr10_-_105604942
|
0.865
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr14_-_23781328
|
0.859
|
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr16_-_66523255
|
0.857
|
NM_001907
|
CTRL
|
chymotrypsin-like
|
|
chr17_+_55854636
|
0.853
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr8_-_145132582
|
0.844
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr12_+_79995927
|
0.837
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr2_+_148495043
|
0.808
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr13_+_112825113
|
0.804
|
NM_000504
|
F10
|
coagulation factor X
|
|
chr20_-_21442663
|
0.803
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr2_+_136005423
|
0.799
|
NM_015361
|
R3HDM1
|
R3H domain containing 1
|
|
chr8_+_123863169
|
0.790
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr2_-_70634129
|
0.785
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr5_+_35653702
|
0.744
|
NM_024867
NM_144722
|
SPEF2
|
sperm flagellar 2
|
|
chr10_+_123862430
|
0.740
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr6_+_32114060
|
0.738
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr2_+_85665041
|
0.735
|
NM_006634
|
VAMP5
|
vesicle-associated membrane protein 5 (myobrevin)
|
|
chr12_+_56406296
|
0.730
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr14_-_34414277
|
0.727
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr13_+_75232797
|
0.725
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr1_-_94779727
|
0.721
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr6_-_52060327
|
0.719
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr16_+_49139694
|
0.715
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr12_-_47604930
|
0.708
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr2_+_201462294
|
0.700
|
NM_001136039
NM_001142355
NM_021824
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr7_+_29200636
|
0.684
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr14_-_23874083
|
0.679
|
NM_001198568
NM_001198592
NM_139247
|
ADCY4
|
adenylate cyclase 4
|
|
chr20_-_22513050
|
0.670
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr20_-_49852353
|
0.670
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr2_+_101981888
|
0.668
|
NM_173343
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr17_+_43165608
|
0.666
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr16_-_87244896
|
0.664
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr2_-_74588293
|
0.657
|
NM_032673
|
PCGF1
|
polycomb group ring finger 1
|
|
chr16_+_65790514
|
0.654
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr6_+_131190237
|
0.648
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr5_-_121440704
|
0.647
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr4_+_114190233
|
0.646
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr20_-_33793507
|
0.635
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr9_+_77695334
|
0.633
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr12_+_94861201
|
0.630
|
NM_152435
|
AMDHD1
|
amidohydrolase domain containing 1
|
|
chr6_-_16869579
|
0.629
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr14_-_34414241
|
0.627
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr14_-_34414201
|
0.626
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr16_-_87535106
|
0.624
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr6_-_138935359
|
0.623
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr2_-_75641535
|
0.620
|
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr3_-_55490442
|
0.619
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr6_+_26153590
|
0.613
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr20_-_22512893
|
0.609
|
|
FOXA2
|
forkhead box A2
|
|
chr19_+_40322231
|
0.607
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr12_+_14409875
|
0.597
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr6_-_128883125
|
0.596
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr20_-_33793591
|
0.589
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr3_-_191522886
|
0.588
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr2_-_197499584
|
0.583
|
NM_024989
|
PGAP1
|
post-GPI attachment to proteins 1
|
|
chr2_-_37727472
|
0.578
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr15_+_97462675
|
0.572
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr19_-_4468715
|
0.571
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr10_-_123343761
|
0.561
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr14_+_20527971
|
0.560
|
|
METTL17
|
methyltransferase like 17
|
|
chr1_-_94475840
|
0.558
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr2_+_201099093
|
0.558
|
NM_001160033
NM_001160046
NM_152524
|
SGOL2
|
shugoshin-like 2 (S. pombe)
|
|
chr7_-_11838260
|
0.556
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr20_-_42249631
|
0.550
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chr2_-_168812132
|
0.542
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr14_-_20642629
|
0.541
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr14_-_22127902
|
0.541
|
|
DAD1
|
defender against cell death 1
|
|
chr17_-_70131389
|
0.541
|
NM_181449
|
CD300E
|
CD300e molecule
|
|
chr3_+_2115449
|
0.535
|
|
CNTN4
|
contactin 4
|
|
chr8_+_121206478
|
0.531
|
NM_021110
|
COL14A1
|
collagen, type XIV, alpha 1
|
|
chr22_+_39404940
|
0.531
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|
|
chr2_-_75641587
|
0.529
|
NM_001135032
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr1_-_94475504
|
0.525
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_+_113061330
|
0.524
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr2_+_96295751
|
0.522
|
|
CIAO1
|
cytosolic iron-sulfur protein assembly 1
|
|
chr14_-_53491019
|
0.521
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr2_+_225973845
|
0.519
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr6_-_41823056
|
0.517
|
NM_001166424
NM_002630
|
PGC
|
progastricsin (pepsinogen C)
|
|
chr9_+_111582317
|
0.514
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr8_+_120497732
|
0.513
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr6_-_149847809
|
0.511
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr10_-_16604009
|
0.511
|
NM_001010908
|
C1QL3
|
complement component 1, q subcomponent-like 3
|
|
chr8_+_85258007
|
0.510
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr14_-_34413831
|
0.506
|
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr8_+_11599119
|
0.506
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr12_-_15265490
|
0.505
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr12_-_6355165
|
0.504
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr3_-_55490401
|
0.503
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr17_+_71154240
|
0.502
|
NM_001162997
|
LOC100130933
|
hypothetical protein LOC100130933
|
|
chr6_+_96570565
|
0.499
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr12_+_51979721
|
0.499
|
NM_021640
|
C12orf10
|
chromosome 12 open reading frame 10
|
|
chr4_+_89597257
|
0.499
|
NM_016323
|
HERC5
|
hect domain and RLD 5
|
|
chr8_-_122722674
|
0.497
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr3_-_81893604
|
0.494
|
NM_000158
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr2_+_238200922
|
0.492
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chrX_+_38305634
|
0.491
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr13_-_110066694
|
0.489
|
|
|
|
|
chr20_+_2309553
|
0.489
|
NM_198994
|
TGM6
|
transglutaminase 6
|
|
chr2_-_96295443
|
0.488
|
|
TMEM127
|
transmembrane protein 127
|
|
chr14_-_34943574
|
0.488
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr5_-_134899537
|
0.486
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr7_-_100751643
|
0.485
|
NM_001130820
NM_001130821
NM_001130822
NM_022777
|
RABL5
|
RAB, member RAS oncogene family-like 5
|
|
chr10_-_71575953
|
0.485
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr10_+_59942874
|
0.485
|
NM_001080512
|
BICC1
|
bicaudal C homolog 1 (Drosophila)
|
|
chr20_-_1922701
|
0.485
|
NM_001190899
NM_001190898
NM_024411
|
PDYN
|
prodynorphin
|
|
chr3_-_121445831
|
0.481
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr2_-_101134146
|
0.481
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr10_+_96295511
|
0.478
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr16_-_67930974
|
0.475
|
NM_032382
|
COG8
|
component of oligomeric golgi complex 8
|
|
chr17_-_40348378
|
0.474
|
NM_001131019
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr14_-_21014871
|
0.474
|
NM_032846
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr4_-_164754106
|
0.472
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr17_+_18796202
|
0.471
|
NM_001042450
NM_152351
|
SLC5A10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10
|
|
chr3_+_113061218
|
0.469
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr17_-_31332568
|
0.469
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr20_-_33793593
|
0.467
|
NM_004902
NM_184234
|
RBM39
|
RNA binding motif protein 39
|
|
chr20_+_43425153
|
0.464
|
NM_001099791
NM_033542
|
SYS1-DBNDD2
SYS1
|
SYS1-DBNDD2 read-through transcript
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
|
|
chr20_-_16501911
|
0.464
|
NM_024704
|
KIF16B
|
kinesin family member 16B
|
|
chr12_+_54361596
|
0.461
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr5_-_35083996
|
0.459
|
NM_031900
|
AGXT2
|
alanine--glyoxylate aminotransferase 2
|
|
chr16_-_80602546
|
0.458
|
NM_145168
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr3_+_112876212
|
0.457
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr16_-_28528780
|
0.456
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr12_-_47605478
|
0.455
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr19_-_53365371
|
0.453
|
NM_000234
|
LIG1
|
ligase I, DNA, ATP-dependent
|
|
chr2_-_201461945
|
0.453
|
NM_032472
NM_130906
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3
|
|
chr2_+_218833982
|
0.452
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr7_+_90177082
|
0.452
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr12_+_54361685
|
0.451
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr14_-_23686269
|
0.450
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr16_-_45735403
|
0.449
|
NM_018092
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr14_+_23633101
|
0.449
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr11_+_12355600
|
0.448
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr11_-_122357545
|
0.447
|
NM_001098169
|
BSX
|
brain-specific homeobox
|
|
chr3_+_113061296
|
0.447
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr22_-_22714230
|
0.447
|
NM_000853
|
GSTT1
|
glutathione S-transferase theta 1
|
|
chr14_+_23700261
|
0.446
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr16_-_45735333
|
0.446
|
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2
|
|
chr2_+_48395271
|
0.446
|
NM_002158
|
FOXN2
|
forkhead box N2
|
|
chr6_-_27387989
|
0.445
|
NM_033482
|
POM121L2
|
POM121 membrane glycoprotein-like 2
|
|
chr3_-_81893308
|
0.440
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr2_-_148495642
|
0.440
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr3_-_185578621
|
0.439
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr18_-_46068048
|
0.439
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr13_+_25726755
|
0.437
|
NM_001260
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr8_+_123863224
|
0.436
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr7_-_50596238
|
0.436
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr9_-_124715392
|
0.435
|
NM_006626
|
ZBTB6
|
zinc finger and BTB domain containing 6
|
|
chr14_+_21015198
|
0.435
|
|
TOX4
|
TOX high mobility group box family member 4
|
|
chr2_-_24123726
|
0.432
|
NM_001142319
NM_025203
|
C2orf44
|
chromosome 2 open reading frame 44
|
|
chr14_+_71469538
|
0.430
|
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr12_+_81604752
|
0.430
|
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr2_-_189752575
|
0.427
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_114947330
|
0.426
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr14_-_21014666
|
0.420
|
NM_001163380
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr6_-_5952096
|
0.419
|
|
NRN1
|
neuritin 1
|
|
chr9_-_26882713
|
0.418
|
NM_001167575
NM_024828
|
C9orf82
|
chromosome 9 open reading frame 82
|
|
chr1_+_107484151
|
0.417
|
NM_001113226
NM_001113228
NM_014917
|
NTNG1
|
netrin G1
|
|
chr4_-_84475328
|
0.416
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr2_-_191723978
|
0.414
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr2_-_68952006
|
0.413
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr2_-_220144427
|
0.413
|
|
OBSL1
|
obscurin-like 1
|
|
chr14_+_21015168
|
0.412
|
NM_014828
|
TOX4
|
TOX high mobility group box family member 4
|
|
chr6_-_149847718
|
0.412
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr10_+_91451313
|
0.410
|
NM_016195
|
KIF20B
|
kinesin family member 20B
|
|
chr3_-_184363126
|
0.409
|
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr4_-_24641404
|
0.408
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr20_+_5839973
|
0.408
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr2_+_190753833
|
0.408
|
NM_001042519
NM_032321
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr2_+_241903395
|
0.407
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr18_-_46068003
|
0.406
|
|
CXXC1
|
CXXC finger protein 1
|
|
chr7_+_96473225
|
0.405
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr2_-_74460933
|
0.405
|
NM_001135040
NM_001190837
NM_004082
|
DCTN1
|
dynactin 1
|
|
chr8_-_99375739
|
0.405
|
NM_024759
|
NIPAL2
|
NIPA-like domain containing 2
|
|
chr7_+_80145461
|
0.405
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_-_81893492
|
0.404
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr14_+_105024576
|
0.402
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr2_+_219141761
|
0.402
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr2_-_74588006
|
0.401
|
|
PCGF1
|
polycomb group ring finger 1
|
|
chr9_-_21984329
|
0.400
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|