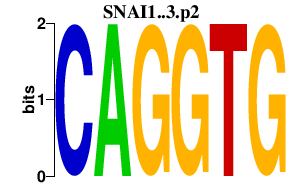
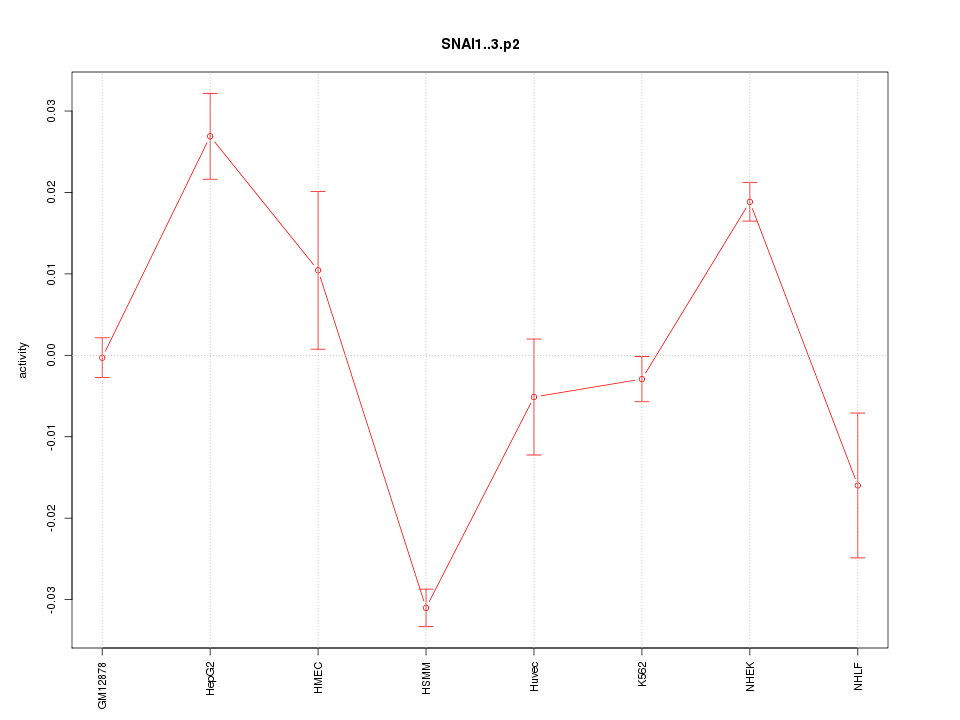
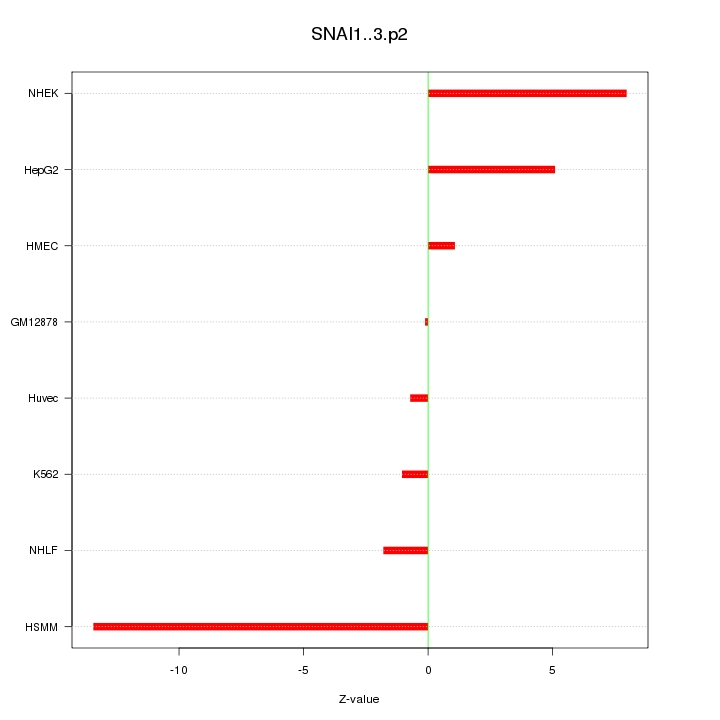
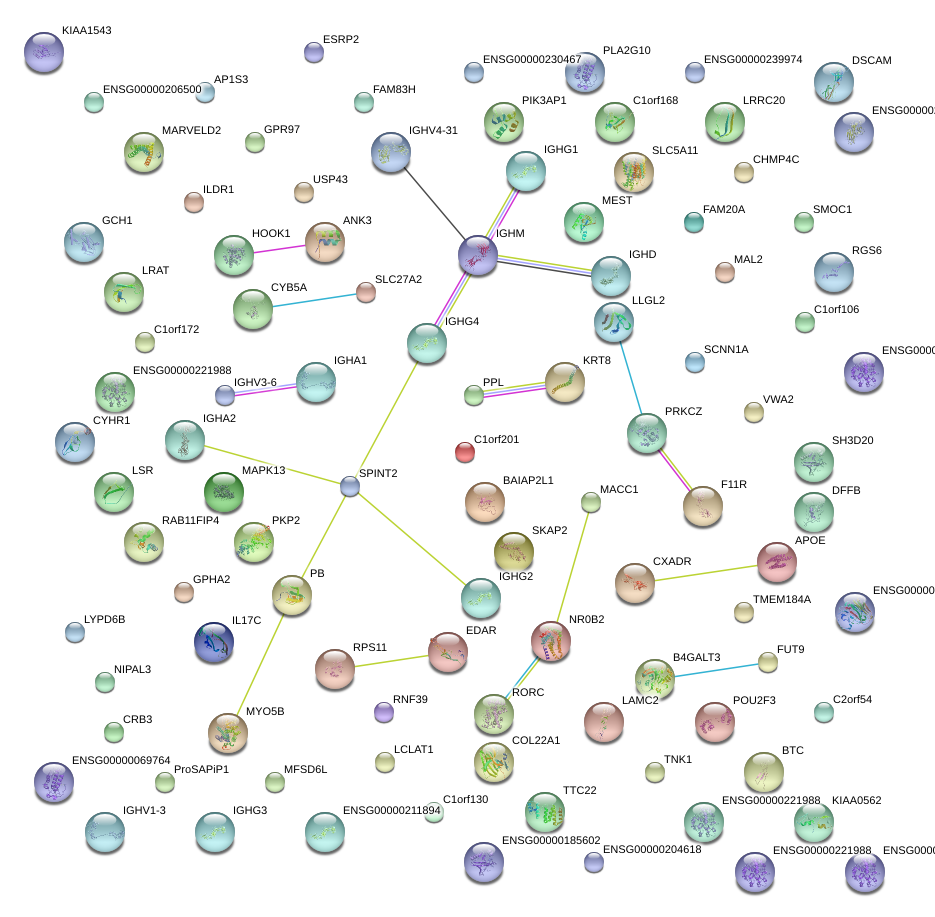
|
chr19_+_40431122
|
11.513
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr1_+_1971762
|
10.834
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr17_+_71033339
|
9.960
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr1_+_60053050
|
9.245
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr14_-_105851580
|
9.214
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr19_+_40431359
|
8.684
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr7_-_1562272
|
8.631
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr2_-_108972100
|
8.049
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr18_-_70110089
|
6.940
|
NM_001190807
NM_001914
NM_148923
|
CYB5A
|
cytochrome b5 type A (microsomal)
|
|
chr12_-_32940931
|
6.799
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr12_-_51585004
|
6.730
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr4_+_155884600
|
6.426
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr2_+_149603226
|
6.148
|
NM_177964
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr19_+_40431618
|
6.023
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr14_+_71468902
|
5.974
|
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr14_-_105948734
|
5.671
|
|
LOC100126583
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_55039458
|
5.568
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr1_+_199130571
|
5.547
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr8_+_82807218
|
5.545
|
NM_152284
|
CHMP4C
|
chromatin modifying protein 4C
|
|
chr8_-_139995417
|
5.431
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr1_-_27113018
|
5.410
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr20_-_3097070
|
5.274
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr1_-_150070849
|
5.236
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr18_-_45975163
|
5.205
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr16_+_87232501
|
5.122
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr4_+_155884564
|
5.078
|
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr10_-_62163083
|
5.042
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr8_-_144887876
|
4.965
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr1_-_159257595
|
4.830
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr6_-_30151526
|
4.787
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr1_-_159257486
|
4.779
|
|
F11R
|
F11 receptor
|
|
chr1_-_159257567
|
4.703
|
|
F11R
|
F11 receptor
|
|
chr17_+_9489578
|
4.577
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr1_+_181259317
|
4.564
|
|
|
|
|
chr1_-_43523825
|
4.519
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr7_-_97868362
|
4.505
|
NM_018842
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr6_+_32230172
|
4.482
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr5_+_68746694
|
4.468
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr15_+_48261649
|
4.381
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr8_-_145661225
|
4.342
|
NM_138496
|
CYHR1
|
cysteine/histidine-rich 1
|
|
chr11_+_119616112
|
4.303
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr2_+_242679507
|
4.302
|
|
LOC728323
|
hypothetical LOC728323
|
|
chr19_+_7566698
|
4.267
|
NM_001080429
NM_020902
|
KIAA1543
|
KIAA1543
|
|
chr15_-_19974402
|
4.219
|
|
|
|
|
chr2_+_30523586
|
4.194
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr1_-_57057956
|
4.185
|
NM_001004303
|
C1orf168
|
chromosome 1 open reading frame 168
|
|
chr10_-_98470164
|
4.179
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr2_-_224410444
|
4.076
|
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr16_-_66827449
|
4.067
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr6_+_96570565
|
4.062
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr17_-_8643163
|
4.048
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr2_-_224410544
|
3.993
|
NM_001039569
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit
|
|
chr17_-_64108529
|
3.990
|
NM_017565
|
FAM20A
|
family with sequence similarity 20, member A
|
|
chr1_+_24614831
|
3.947
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr16_+_56259657
|
3.926
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr1_-_27159462
|
3.926
|
NM_152365
|
C1orf172
|
chromosome 1 open reading frame 172
|
|
chr14_-_105845570
|
3.899
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr17_+_26839143
|
3.894
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr14_-_105900687
|
3.869
|
|
IGHA1
IGHD
IGHG3
IGHM
SKAP2
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
src kinase associated phosphoprotein 2
|
|
chr19_+_6415259
|
3.854
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr16_-_4927057
|
3.840
|
NM_002705
|
PPL
|
periplakin
|
|
chr2_-_241484245
|
3.834
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr1_-_24614173
|
3.813
|
NM_001199012
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr21_+_17807167
|
3.813
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_-_159413833
|
3.811
|
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr8_+_120289735
|
3.782
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr7_-_20223467
|
3.732
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr21_-_41140908
|
3.720
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr19_+_43447262
|
3.717
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_+_24755152
|
3.698
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr11_-_64459910
|
3.687
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr6_+_36206239
|
3.651
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr10_-_71753684
|
3.650
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr12_-_6355165
|
3.620
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr7_+_129913271
|
3.616
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr16_+_24765052
|
3.577
|
NM_052944
|
SLC5A11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11
|
|
chr17_+_7225088
|
3.555
|
NM_003985
|
TNK1
|
tyrosine kinase, non-receptor, 1
|
|
chr1_+_181421984
|
3.524
|
|
LAMC2
|
laminin, gamma 2
|
|
chr3_-_123223658
|
3.505
|
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr22_+_21027538
|
3.494
|
|
|
|
|
chr4_-_75938710
|
3.439
|
NM_001729
|
BTC
|
betacellulin
|
|
chr1_-_3763480
|
3.433
|
|
KIAA0562
|
KIAA0562
|
|
chr2_-_108972048
|
3.411
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr14_+_69415880
|
3.410
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr14_-_54438913
|
3.401
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_-_40858775
|
3.383
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr14_-_106270497
|
3.379
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr1_+_181422016
|
3.269
|
|
LAMC2
|
laminin, gamma 2
|
|
chr10_+_115988999
|
3.251
|
NM_198496
|
VWA2
|
von Willebrand factor A domain containing 2
|
|
chr7_-_97868101
|
3.212
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr16_-_14695930
|
3.208
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr1_-_3763620
|
3.186
|
NM_014704
|
KIAA0562
|
KIAA0562
|
|
chr1_+_181421796
|
3.177
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_3763704
|
3.136
|
NM_004402
|
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase)
|
|
chr11_+_120828058
|
3.089
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_+_95355415
|
3.037
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr17_-_37034288
|
3.035
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr9_-_138757175
|
3.022
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr14_-_105549177
|
3.017
|
|
IGHV4-31
IGHD
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr11_+_120828275
|
3.010
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chrX_-_153256122
|
3.004
|
|
FLNA
|
filamin A, alpha
|
|
chr5_-_101659864
|
3.003
|
NM_180991
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1
|
|
chr16_+_23101538
|
3.003
|
NM_001039
|
SCNN1G
|
sodium channel, nonvoltage-gated 1, gamma
|
|
chr16_+_70217564
|
3.000
|
NM_001017967
NM_052858
|
MARVELD3
|
MARVEL domain containing 3
|
|
chr20_+_2744947
|
2.942
|
NM_001167670
|
LOC100288797
|
hypothetical protein LOC100288797
|
|
chr8_-_75396016
|
2.934
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr12_-_6354818
|
2.930
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr14_-_106202262
|
2.930
|
|
IGLJ3
IGHA1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
|
|
chr20_-_49592654
|
2.894
|
NM_012340
NM_173091
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr17_-_64108331
|
2.858
|
|
FAM20A
|
family with sequence similarity 20, member A
|
|
chr19_-_8670054
|
2.856
|
NM_178525
|
ACTL9
|
actin-like 9
|
|
chr16_+_88280569
|
2.839
|
NM_001098533
NM_001160367
NM_052987
NM_052988
|
CDK10
|
cyclin-dependent kinase 10
|
|
chr7_+_29570024
|
2.833
|
|
PRR15
|
proline rich 15
|
|
chr16_-_28843955
|
2.825
|
NM_024816
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2
|
|
chr6_+_33280396
|
2.808
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr6_+_32229771
|
2.807
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr19_-_3928516
|
2.805
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr20_+_43531806
|
2.799
|
NM_006103
|
WFDC2
|
WAP four-disulfide core domain 2
|
|
chr11_+_27019084
|
2.797
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr14_-_106084085
|
2.756
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_+_149612488
|
2.728
|
NM_003828
|
MTMR1
|
myotubularin related protein 1
|
|
chr1_-_229181219
|
2.706
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr19_-_53914768
|
2.689
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_+_229181462
|
2.684
|
|
ARV1
|
ARV1 homolog (S. cerevisiae)
|
|
chr14_-_105644300
|
2.680
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_229181229
|
2.655
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr22_+_18124225
|
2.652
|
NM_005992
NM_080646
NM_080647
|
TBX1
|
T-box 1
|
|
chr11_+_128751044
|
2.648
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr17_-_24253821
|
2.633
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr1_+_229181417
|
2.622
|
NM_022786
|
ARV1
|
ARV1 homolog (S. cerevisiae)
|
|
chr8_+_102573837
|
2.594
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_-_149955415
|
2.589
|
NM_001172648
NM_001172649
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr3_-_69064364
|
2.586
|
NM_001005527
NM_182522
|
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4
|
|
chr13_+_112671497
|
2.582
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr5_+_68824143
|
2.580
|
|
OCLN
|
occludin
|
|
chr14_-_23106372
|
2.578
|
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr14_-_105589467
|
2.573
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_96167900
|
2.546
|
NM_001002036
|
ASTL
|
astacin-like metallo-endopeptidase (M12 family)
|
|
chr9_+_107046811
|
2.545
|
|
SLC44A1
|
solute carrier family 44, member 1
|
|
chr19_+_43447078
|
2.543
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_229181240
|
2.534
|
NM_001122835
NM_024525
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr16_+_88280594
|
2.532
|
|
CDK10
|
cyclin-dependent kinase 10
|
|
chr1_-_229181203
|
2.531
|
|
TTC13
|
tetratricopeptide repeat domain 13
|
|
chr17_-_7047285
|
2.528
|
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr14_+_104852225
|
2.511
|
|
PACS2
|
phosphofurin acidic cluster sorting protein 2
|
|
chr19_-_59368625
|
2.489
|
NM_001145303
NM_144686
|
TMC4
|
transmembrane channel-like 4
|
|
chr5_-_60175798
|
2.486
|
NM_001104558
NM_024930
|
ELOVL7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast)
|
|
chr3_-_49434874
|
2.469
|
|
AMT
|
aminomethyltransferase
|
|
chr9_-_125070675
|
2.458
|
NM_001171137
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr1_-_199635057
|
2.454
|
|
LAD1
|
ladinin 1
|
|
chr4_-_35922288
|
2.448
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr8_+_145692916
|
2.440
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr17_+_24944605
|
2.394
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr8_-_79880095
|
2.378
|
NM_000880
|
IL7
|
interleukin 7
|
|
chr20_-_20641122
|
2.369
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr16_+_2016807
|
2.366
|
NM_001130012
NM_004785
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr2_+_218992053
|
2.361
|
NM_007127
|
VIL1
|
villin 1
|
|
chr17_+_38064787
|
2.359
|
NM_016437
|
TUBG2
|
tubulin, gamma 2
|
|
chr17_+_2627099
|
2.356
|
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr9_-_109290528
|
2.348
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_102574012
|
2.339
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr3_+_39826032
|
2.337
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr7_-_22363036
|
2.323
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr6_+_34964999
|
2.316
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr17_-_37196463
|
2.313
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr16_-_30477006
|
2.290
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr15_+_92575767
|
2.277
|
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr11_+_45900731
|
2.249
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr11_-_129322510
|
2.237
|
NM_199438
NM_199439
|
PRDM10
|
PR domain containing 10
|
|
chr3_-_27739201
|
2.233
|
|
EOMES
|
eomesodermin
|
|
chr2_+_231610438
|
2.205
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr14_-_105804211
|
2.197
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr18_+_32021418
|
2.195
|
NM_017947
|
MOCOS
|
molybdenum cofactor sulfurase
|
|
chr2_-_101134146
|
2.175
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr12_+_6926014
|
2.149
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr10_-_82039158
|
2.147
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr16_+_67236730
|
2.145
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_-_53784494
|
2.144
|
NM_003168
|
SUPT4H1
|
suppressor of Ty 4 homolog 1 (S. cerevisiae)
|
|
chr4_-_54152480
|
2.132
|
NM_001126328
|
LNX1
|
ligand of numb-protein X 1
|
|
chr2_-_166359016
|
2.124
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr18_-_42353031
|
2.112
|
NM_001145473
NM_001173129
|
LOXHD1
|
lipoxygenase homology domains 1
|
|
chr11_+_111294868
|
2.108
|
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr11_+_111294810
|
2.106
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr20_-_44612619
|
2.103
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr1_+_3360880
|
2.097
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr4_-_40327396
|
2.091
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr16_-_10183244
|
2.077
|
NM_001134408
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr20_+_18066473
|
2.068
|
NM_001164811
|
PET117
|
cytochrome c oxidase assembly factor-like
|
|
chr16_+_49139694
|
2.066
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr20_+_36663865
|
2.065
|
NM_001164431
|
ARHGAP40
|
Rho GTPase activating protein 40
|
|
chr12_-_30740011
|
2.059
|
|
IPO8
|
importin 8
|
|
chr4_-_11039600
|
2.054
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr22_+_23329113
|
2.048
|
NM_001032364
NM_005265
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr6_+_31903374
|
2.048
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr22_-_35835548
|
2.046
|
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr17_-_33982580
|
2.040
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr17_-_64108843
|
2.039
|
|
FAM20A
|
family with sequence similarity 20, member A
|
|
chr13_-_31784088
|
2.034
|
NM_001136571
|
ZAR1L
|
zygote arrest 1-like
|
|
chr20_-_22514092
|
2.029
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr2_-_70871239
|
2.028
|
NM_001004311
|
FIGLA
|
folliculogenesis specific basic helix-loop-helix
|