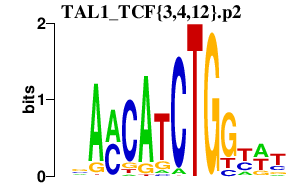
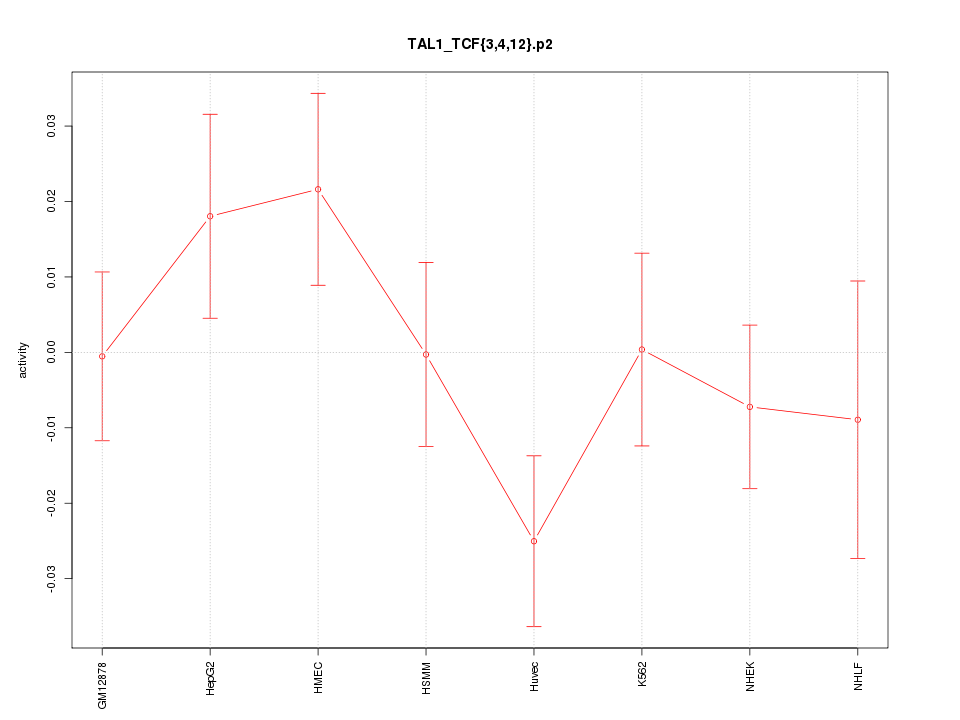
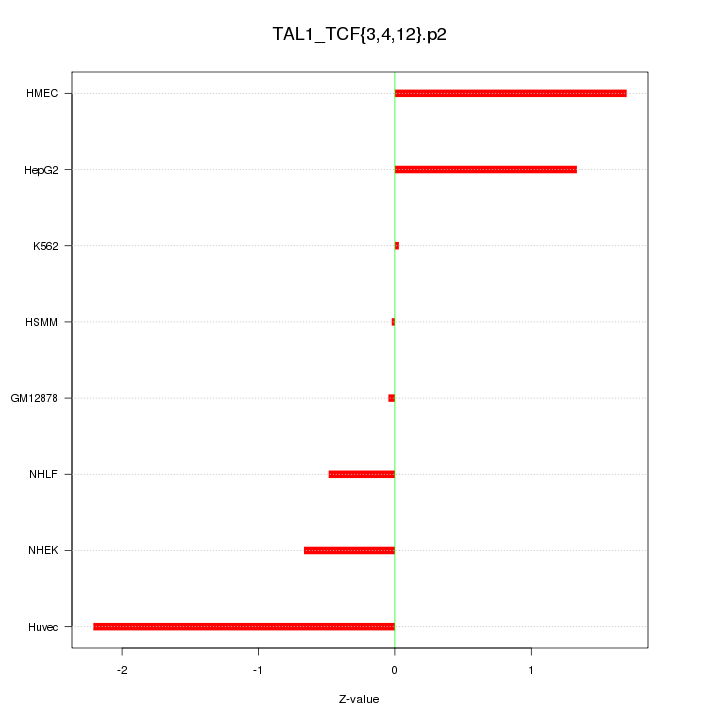
|
chr4_-_69116839
|
1.443
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr6_-_29431950
|
1.168
|
NM_030876
|
OR5V1
|
olfactory receptor, family 5, subfamily V, member 1
|
|
chr1_+_50344550
|
1.051
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_205272619
|
0.980
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr2_+_151922350
|
0.971
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr2_-_96320185
|
0.933
|
|
SNRNP200
|
small nuclear ribonucleoprotein 200kDa (U5)
|
|
chr13_-_85271329
|
0.920
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr10_+_5228756
|
0.910
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr20_+_36643251
|
0.840
|
NM_001018082
|
ADIG
|
adipogenin
|
|
chr5_+_167651063
|
0.824
|
NM_001161661
NM_001161662
NM_015238
|
WWC1
|
WW and C2 domain containing 1
|
|
chr5_+_147423727
|
0.766
|
NM_001127698
NM_001127699
NM_006846
|
SPINK5
|
serine peptidase inhibitor, Kazal type 5
|
|
chr2_-_133256097
|
0.756
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr5_+_173405343
|
0.755
|
|
HMP19
|
HMP19 protein
|
|
chr8_-_68821131
|
0.696
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr11_-_4827013
|
0.694
|
NM_001004758
|
OR51S1
|
olfactory receptor, family 51, subfamily S, member 1
|
|
chr5_+_173405298
|
0.630
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr17_+_38256886
|
0.619
|
|
|
|
|
chr22_+_23309717
|
0.609
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr16_-_29818334
|
0.609
|
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr16_-_29817841
|
0.585
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr6_-_66473790
|
0.583
|
NM_001142800
NM_001142801
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr9_-_35860316
|
0.581
|
NM_001004487
|
OR13J1
|
olfactory receptor, family 13, subfamily J, member 1
|
|
chr21_+_17841297
|
0.564
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chrX_+_46967360
|
0.564
|
NM_001170460
|
CDK16
|
cyclin-dependent kinase 16
|
|
chr2_+_191009265
|
0.554
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr1_+_156842852
|
0.545
|
NM_001004478
|
OR10Z1
|
olfactory receptor, family 10, subfamily Z, member 1
|
|
chr9_+_114376709
|
0.515
|
|
KIAA1958
|
KIAA1958
|
|
chr6_+_126282078
|
0.507
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr2_+_187173176
|
0.500
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr10_-_75127559
|
0.496
|
NM_001144000
|
AGAP5
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 5
|
|
chr6_-_29451042
|
0.494
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr1_+_246718487
|
0.493
|
NM_001004697
|
OR2T5
|
olfactory receptor, family 2, subfamily T, member 5
|
|
chr1_+_19807168
|
0.486
|
|
|
|
|
chr2_-_225519974
|
0.486
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr4_-_152368492
|
0.482
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr2_-_233586145
|
0.478
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr4_+_73123712
|
0.474
|
NM_001144756
NM_053036
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr22_+_43451600
|
0.466
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr11_-_88436463
|
0.454
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr3_-_10307408
|
0.439
|
NM_001134941
NM_016362
|
GHRL
|
ghrelin/obestatin prepropeptide
|
|
chr9_+_6318348
|
0.435
|
NM_001001874
NM_001001875
NM_033516
|
TPD52L3
|
tumor protein D52-like 3
|
|
chr22_-_14829803
|
0.418
|
NM_001005239
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1
|
|
chr3_-_196792059
|
0.409
|
|
APOD
|
apolipoprotein D
|
|
chr18_+_19826734
|
0.394
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr1_+_32489426
|
0.386
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr3_+_134976965
|
0.386
|
|
TF
|
transferrin
|
|
chr1_-_169888373
|
0.382
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr12_-_101835183
|
0.372
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr1_-_203657803
|
0.371
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr16_+_56286212
|
0.361
|
NM_032269
|
CCDC135
|
coiled-coil domain containing 135
|
|
chr12_+_63510481
|
0.359
|
|
|
|
|
chr14_+_19513442
|
0.357
|
NM_001005486
|
OR4K15
|
olfactory receptor, family 4, subfamily K, member 15
|
|
chr12_-_48508411
|
0.356
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr17_-_36409632
|
0.348
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr19_+_15765760
|
0.344
|
NM_001004466
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5
|
|
chr2_+_232979795
|
0.342
|
NM_031313
|
ALPPL2
|
alkaline phosphatase, placental-like 2
|
|
chr3_-_191650291
|
0.338
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr11_-_5212168
|
0.338
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr11_+_5558682
|
0.331
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr1_-_246789396
|
0.324
|
NM_001004694
|
OR2T29
OR2T5
|
olfactory receptor, family 2, subfamily T, member 29
olfactory receptor, family 2, subfamily T, member 5
|
|
chr12_-_46405477
|
0.319
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr7_+_31693155
|
0.319
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr7_+_94953148
|
0.318
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr14_-_93919305
|
0.309
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr12_-_101835026
|
0.306
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr14_+_19598043
|
0.305
|
NM_001004717
|
OR4L1
|
olfactory receptor, family 4, subfamily L, member 1
|
|
chr11_-_62445308
|
0.304
|
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr19_+_61624
|
0.304
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr10_+_85923473
|
0.298
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chr16_+_4546491
|
0.294
|
NM_001145011
|
LOC342346
|
hypothetical protein LOC342346
|
|
chrX_-_15593074
|
0.292
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr6_-_41230003
|
0.291
|
NM_178174
|
TREML1
|
triggering receptor expressed on myeloid cells-like 1
|
|
chr13_-_26232777
|
0.291
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr17_-_53761119
|
0.287
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr7_-_41709191
|
0.286
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr8_+_110443881
|
0.283
|
NM_177531
|
PKHD1L1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1
|
|
chr2_+_108360798
|
0.283
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr14_-_93493519
|
0.282
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr3_-_196792274
|
0.280
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr9_+_124355211
|
0.273
|
NM_001004457
|
OR1N2
|
olfactory receptor, family 1, subfamily N, member 2
|
|
chr12_+_115481568
|
0.272
|
NM_001085481
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr1_+_179164195
|
0.272
|
|
|
|
|
chr14_+_19735334
|
0.259
|
NM_001005503
|
OR11G2
|
olfactory receptor, family 11, subfamily G, member 2
|
|
chr12_-_101835510
|
0.258
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr8_-_20084892
|
0.258
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr17_+_40594046
|
0.256
|
NM_144608
|
HEXIM2
|
hexamthylene bis-acetamide inducible 2
|
|
chr2_-_233586164
|
0.246
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr6_-_30236667
|
0.241
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr3_+_73193499
|
0.238
|
NM_018029
|
EBLN2
|
endogenous Borna-like N element-2
|
|
chr8_+_54926920
|
0.237
|
NM_170587
|
RGS20
|
regulator of G-protein signaling 20
|
|
chr16_+_21623784
|
0.237
|
NM_170664
|
OTOA
|
otoancorin
|
|
chr1_+_54786394
|
0.237
|
NM_015547
NM_147161
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr18_+_30589937
|
0.236
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr7_+_75977680
|
0.233
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr5_-_56814310
|
0.231
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr9_+_35663852
|
0.231
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr15_-_40537002
|
0.226
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr11_-_71424614
|
0.225
|
|
|
|
|
chr6_-_42798286
|
0.225
|
NM_000322
|
PRPH2
|
peripherin 2 (retinal degeneration, slow)
|
|
chr6_+_163068991
|
0.224
|
NM_001080379
|
PACRG
|
PARK2 co-regulated
|
|
chr2_-_166638376
|
0.223
|
NM_001165963
NM_001165964
NM_006920
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr12_+_61240542
|
0.219
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr6_-_169392771
|
0.218
|
|
|
|
|
chr6_+_29472310
|
0.214
|
NM_013936
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2
|
|
chr11_+_5367182
|
0.213
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr9_+_106371269
|
0.211
|
NM_001004483
|
OR13C8
|
olfactory receptor, family 13, subfamily C, member 8
|
|
chr22_-_41672990
|
0.210
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr19_+_17499064
|
0.209
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr17_-_59342319
|
0.206
|
NM_001318
NM_022579
NM_022580
NM_022581
|
CSHL1
|
chorionic somatomammotropin hormone-like 1
|
|
chr10_+_44726635
|
0.203
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr21_-_33107813
|
0.200
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr20_-_44612619
|
0.199
|
NM_080721
|
C20orf123
|
chromosome 20 open reading frame 123
|
|
chr2_-_240618518
|
0.194
|
NM_001005853
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2
|
|
chr17_-_10362583
|
0.194
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr11_-_56166490
|
0.190
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr21_-_26345294
|
0.190
|
|
|
|
|
chr1_+_246703249
|
0.188
|
NM_001005495
|
OR2T3
|
olfactory receptor, family 2, subfamily T, member 3
|
|
chr18_-_55136673
|
0.184
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr17_+_35372751
|
0.183
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr10_+_71482362
|
0.182
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr11_-_74478405
|
0.182
|
NM_001005285
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4
|
|
chr1_+_50342168
|
0.181
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_-_154043403
|
0.179
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr6_-_11490487
|
0.178
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr9_-_106407728
|
0.178
|
NM_001004481
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2
|
|
chr1_-_6343306
|
0.178
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr3_+_196933481
|
0.175
|
|
MUC20
|
mucin 20, cell surface associated
|
|
chr11_+_66979393
|
0.174
|
NM_145200
|
CABP4
|
calcium binding protein 4
|
|
chr17_+_36636404
|
0.173
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr14_+_38804226
|
0.173
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr10_-_50012058
|
0.172
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr10_-_14090459
|
0.171
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr5_-_131907071
|
0.171
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr1_+_24713396
|
0.169
|
|
RCAN3
|
RCAN family member 3
|
|
chr1_-_156800017
|
0.169
|
NM_001160325
|
OR6P1
|
olfactory receptor, family 6, subfamily P, member 1
|
|
chr11_-_66431910
|
0.166
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr1_+_245835478
|
0.164
|
NM_001001914
|
OR2G3
|
olfactory receptor, family 2, subfamily G, member 3
|
|
chr16_-_20246309
|
0.164
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr19_+_58239802
|
0.160
|
NM_001191055
|
HERV-V2
|
envelope glycoprotein ENVV2
|
|
chr7_+_141124710
|
0.159
|
NM_016944
|
TAS2R4
|
taste receptor, type 2, member 4
|
|
chr10_-_98311813
|
0.158
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr9_-_28709302
|
0.156
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr5_-_88235624
|
0.155
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr22_-_34930705
|
0.154
|
NM_030643
NM_145660
|
APOL4
|
apolipoprotein L, 4
|
|
chr11_-_65182411
|
0.153
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr2_-_65013070
|
0.152
|
|
LOC400958
|
hypothetical LOC400958
|
|
chr11_+_6085489
|
0.151
|
NM_001005181
|
OR56B4
|
olfactory receptor, family 56, subfamily B, member 4
|
|
chr3_+_23904086
|
0.151
|
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast)
|
|
chr6_-_150253729
|
0.149
|
NM_139165
|
RAET1E
|
retinoic acid early transcript 1E
|
|
chr1_+_221168388
|
0.149
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chrX_-_110400406
|
0.147
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr11_-_62445587
|
0.145
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr10_-_121286034
|
0.144
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr8_+_21967008
|
0.143
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr12_+_118590039
|
0.143
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr3_-_121550767
|
0.142
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr1_-_24179326
|
0.141
|
|
SRSF10
|
serine/arginine-rich splicing factor 10
|
|
chr17_-_77396372
|
0.139
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr14_+_19414230
|
0.139
|
NM_001005501
|
OR4K2
|
olfactory receptor, family 4, subfamily K, member 2
|
|
chr19_-_8473473
|
0.137
|
NM_032152
|
PRAM1
|
PML-RARA regulated adaptor molecule 1
|
|
chr7_-_141187689
|
0.137
|
NM_001008270
NM_001171951
|
PRSS37
|
protease, serine, 37
|
|
chr1_-_246554492
|
0.136
|
NM_001004691
|
OR2M7
|
olfactory receptor, family 2, subfamily M, member 7
|
|
chrX_-_33267646
|
0.136
|
NM_000109
|
DMD
|
dystrophin
|
|
chr9_-_106420305
|
0.133
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr1_+_28134090
|
0.133
|
NM_001009568
NM_014474
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr10_+_85994788
|
0.133
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chrX_+_77267477
|
0.132
|
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr5_-_135318420
|
0.132
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr1_-_49014993
|
0.131
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr17_-_35911340
|
0.131
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr5_+_134331494
|
0.131
|
NM_178019
|
CATSPER3
|
cation channel, sperm associated 3
|
|
chr20_-_39199936
|
0.130
|
|
|
|
|
chr7_-_143560850
|
0.129
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr11_-_123130435
|
0.128
|
NM_001005188
|
OR6X1
|
olfactory receptor, family 6, subfamily X, member 1
|
|
chr3_+_120982020
|
0.128
|
NM_003889
NM_033013
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr15_+_56511466
|
0.125
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr7_-_99365178
|
0.124
|
NM_181538
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa
|
|
chr1_-_158948176
|
0.123
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr2_-_97775300
|
0.121
|
|
TMEM131
|
transmembrane protein 131
|
|
chr3_+_142525744
|
0.119
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr14_+_18447521
|
0.119
|
NM_001013354
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12
|
|
chr16_+_66836798
|
0.118
|
|
PLA2G15
|
phospholipase A2, group XV
|
|
chr7_+_155129943
|
0.118
|
NM_053043
|
RBM33
|
RNA binding motif protein 33
|
|
chr7_+_90176647
|
0.116
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr1_+_156991133
|
0.113
|
NM_001005184
|
OR6K6
|
olfactory receptor, family 6, subfamily K, member 6
|
|
chr12_-_121753795
|
0.113
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr10_-_105410840
|
0.112
|
|
|
|
|
chrX_-_55037235
|
0.109
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr11_-_4782185
|
0.109
|
NM_001005177
|
OR52R1
|
olfactory receptor, family 52, subfamily R, member 1
|
|
chr10_-_921694
|
0.108
|
NM_015155
|
LARP4B
|
La ribonucleoprotein domain family, member 4B
|
|
chr18_+_50409387
|
0.108
|
NM_173629
|
C18orf26
|
chromosome 18 open reading frame 26
|
|
chr22_+_20880112
|
0.108
|
|
|
|
|
chr6_+_36318922
|
0.108
|
NM_173676
NM_001145716
|
PNPLA1
|
patatin-like phospholipase domain containing 1
|
|
chr21_+_44844924
|
0.108
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr19_-_51319769
|
0.108
|
NM_207393
|
IGFL3
|
IGF-like family member 3
|
|
chr2_+_73911477
|
0.105
|
|
STAMBP
|
STAM binding protein
|
|
chr11_+_72658586
|
0.104
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr2_-_157892391
|
0.102
|
NM_001009959
|
ERMN
|
ermin, ERM-like protein
|
|
chr7_+_150013716
|
0.102
|
NM_015660
|
GIMAP2
|
GTPase, IMAP family member 2
|
|
chr6_+_167456230
|
0.100
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|