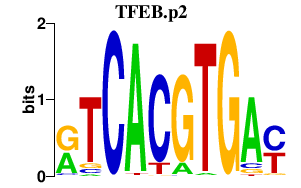
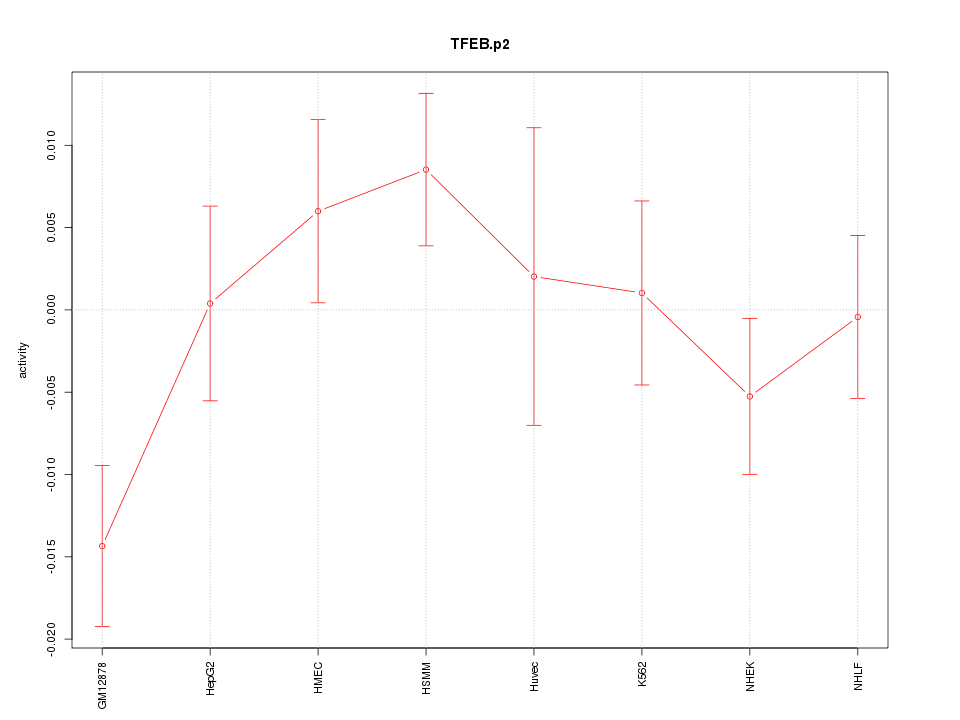
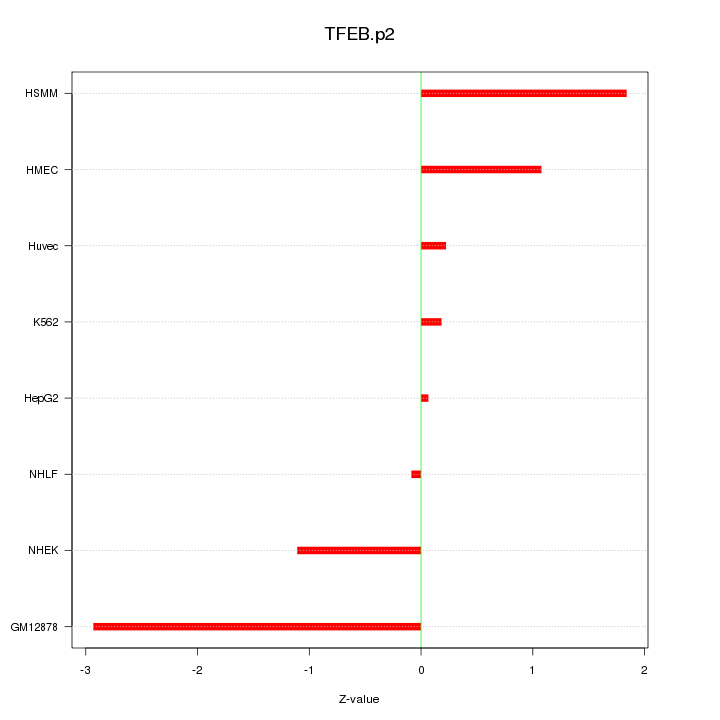
|
chr20_+_43953428
|
1.710
|
|
CTSA
|
cathepsin A
|
|
chr6_-_84994032
|
1.544
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr20_-_43953036
|
1.502
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr1_-_68470585
|
1.493
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr19_+_10625987
|
1.485
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr10_-_32257693
|
1.475
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr13_+_112999538
|
1.361
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr20_+_43953606
|
1.343
|
|
CTSA
|
cathepsin A
|
|
chr1_-_11788550
|
1.332
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr17_-_7078446
|
1.329
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_-_10625449
|
1.328
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr13_+_112999590
|
1.309
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_48582389
|
1.264
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr16_+_28893525
|
1.264
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr16_+_68542126
|
1.212
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr2_+_176689737
|
1.202
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr5_+_140200995
|
1.128
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr17_-_7078304
|
1.121
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr17_-_7078399
|
1.108
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr1_-_202595666
|
1.064
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr14_-_19999475
|
1.064
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr1_-_152459506
|
1.033
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr10_-_32257768
|
1.030
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr13_+_112999462
|
1.023
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr1_-_152459667
|
1.016
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr1_+_201364050
|
1.013
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr11_-_85457469
|
1.003
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_+_130749791
|
0.964
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr1_-_11788666
|
0.964
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr8_+_71744153
|
0.949
|
NM_001011720
|
XKR9
|
XK, Kell blood group complex subunit-related family, member 9
|
|
chr20_+_43953372
|
0.928
|
|
CTSA
|
cathepsin A
|
|
chr4_-_164473107
|
0.926
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr7_-_47588652
|
0.924
|
|
TNS3
|
tensin 3
|
|
chr9_-_130749604
|
0.911
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr11_-_85457736
|
0.904
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr8_+_98857190
|
0.892
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr11_-_85457753
|
0.888
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_+_130749812
|
0.876
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr5_+_150807331
|
0.871
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr1_-_152797767
|
0.870
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr14_-_91642703
|
0.863
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr18_+_9126742
|
0.851
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr6_-_109810396
|
0.842
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr1_-_39097904
|
0.835
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr5_+_150807380
|
0.829
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr19_+_10625896
|
0.828
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr3_-_127558747
|
0.815
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr9_-_34038869
|
0.809
|
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr1_-_94475840
|
0.809
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_159550789
|
0.807
|
NM_001035511
NM_001035512
NM_001035513
NM_003001
|
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa
|
|
chr16_+_68542310
|
0.799
|
NM_182619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr4_+_128773529
|
0.793
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr20_-_32877029
|
0.791
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr1_-_152797820
|
0.789
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr8_-_68103339
|
0.789
|
NM_001013626
|
LRRC67
|
leucine rich repeat containing 67
|
|
chr8_+_133856738
|
0.784
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr4_-_100461530
|
0.778
|
NM_000668
|
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr8_+_76059262
|
0.769
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr14_-_91642641
|
0.757
|
|
ATXN3
ATXN8
|
ataxin 3
ataxin 8
|
|
chrX_+_54483565
|
0.757
|
NM_058163
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr6_+_87921980
|
0.740
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr11_-_47427263
|
0.735
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr20_-_61317891
|
0.723
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr15_-_48766252
|
0.721
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr16_+_30952350
|
0.719
|
|
STX4
|
syntaxin 4
|
|
chr2_+_176761552
|
0.719
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr8_+_133856793
|
0.714
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr9_-_34038887
|
0.712
|
NM_018449
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr8_-_54918099
|
0.706
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr6_-_80713589
|
0.706
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr2_+_27659396
|
0.700
|
NM_032434
|
ZNF512
|
zinc finger protein 512
|
|
chr20_+_37024383
|
0.695
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr5_-_131160541
|
0.693
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr11_-_71491916
|
0.686
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr3_-_171786468
|
0.681
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr3_-_172109075
|
0.678
|
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr1_-_152797760
|
0.672
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_+_185515013
|
0.672
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr15_+_46200460
|
0.667
|
NM_205850
|
SLC24A5
|
solute carrier family 24, member 5
|
|
chr6_+_127629677
|
0.667
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr12_-_56432385
|
0.666
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chrX_+_149487677
|
0.664
|
NM_000252
|
MTM1
|
myotubularin 1
|
|
chr17_-_64462976
|
0.663
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr9_-_96395828
|
0.662
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr14_-_54025414
|
0.659
|
|
GMFB
|
glia maturation factor, beta
|
|
chr14_-_54025464
|
0.651
|
NM_004124
|
GMFB
|
glia maturation factor, beta
|
|
chr13_-_109236897
|
0.649
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr12_+_52713098
|
0.646
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr15_-_48766213
|
0.646
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr16_+_30952382
|
0.642
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr3_+_186012670
|
0.639
|
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae)
|
|
chr3_-_172109119
|
0.626
|
NM_020390
|
EIF5A2
|
eukaryotic translation initiation factor 5A2
|
|
chr16_+_87450991
|
0.625
|
NM_016209
|
TRAPPC2L
|
trafficking protein particle complex 2-like
|
|
chr11_+_67532592
|
0.617
|
NM_001161473
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1
|
|
chr1_-_239587090
|
0.611
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr4_+_128870968
|
0.608
|
NM_031291
|
SLC25A31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chrX_+_149487741
|
0.608
|
|
MTM1
|
myotubularin 1
|
|
chr16_-_1465041
|
0.603
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr7_+_150390566
|
0.602
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr1_+_173235193
|
0.593
|
NM_001007214
|
CACYBP
|
calcyclin binding protein
|
|
chr2_-_133144094
|
0.592
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr1_-_94475504
|
0.591
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr11_+_9642226
|
0.586
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr11_+_9642229
|
0.582
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr5_-_141372721
|
0.581
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr10_+_70553936
|
0.579
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr15_-_48766275
|
0.579
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr6_-_163754903
|
0.578
|
|
LOC100526820
|
hypothetical LOC100526820
|
|
chr2_-_39859912
|
0.574
|
NM_025264
|
THUMPD2
|
THUMP domain containing 2
|
|
chr6_-_109810246
|
0.574
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr11_-_71491942
|
0.570
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr11_+_9642200
|
0.569
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr2_+_26249463
|
0.568
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr7_-_158304062
|
0.566
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr1_-_153195188
|
0.565
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr14_-_19999324
|
0.561
|
|
TMEM55B
|
transmembrane protein 55B
|
|
chrX_+_101862297
|
0.560
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr5_+_150807397
|
0.556
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr19_+_45948598
|
0.553
|
NM_004596
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr7_-_13997389
|
0.549
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr15_-_71712711
|
0.546
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr3_-_158700036
|
0.542
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr5_+_34692239
|
0.542
|
|
RAI14
|
retinoic acid induced 14
|
|
chr8_-_71743973
|
0.541
|
NM_016027
|
LACTB2
|
lactamase, beta 2
|
|
chr16_-_1465021
|
0.540
|
|
CLCN7
|
chloride channel 7
|
|
chr15_-_71712605
|
0.538
|
|
NPTN
|
neuroplastin
|
|
chr6_-_33822659
|
0.535
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr5_+_140194152
|
0.531
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr1_-_152459697
|
0.531
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr19_-_13088212
|
0.530
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr13_-_49057629
|
0.530
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr7_+_32501665
|
0.527
|
NM_015060
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr12_-_122415708
|
0.523
|
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr6_+_123152044
|
0.518
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr4_+_56414565
|
0.517
|
NM_001024924
NM_018261
NM_178237
|
EXOC1
|
exocyst complex component 1
|
|
chr10_+_51497660
|
0.516
|
NM_001005751
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr18_+_75967902
|
0.514
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr13_-_35818603
|
0.514
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr5_+_52812236
|
0.509
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr2_-_27399405
|
0.509
|
NM_002437
|
MPV17
|
MpV17 mitochondrial inner membrane protein
|
|
chr19_-_3936460
|
0.508
|
NM_001961
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr17_-_68769613
|
0.507
|
NM_001129885
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like
|
|
chr1_-_26105477
|
0.507
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr20_-_60484420
|
0.506
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr11_+_85017269
|
0.503
|
NM_001193537
NM_001193538
NM_018480
|
TMEM126B
|
transmembrane protein 126B
|
|
chr2_+_198073358
|
0.502
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr1_-_239587007
|
0.498
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr1_-_153195219
|
0.497
|
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr3_-_121445831
|
0.493
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr11_+_9642182
|
0.491
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr3_+_134601479
|
0.487
|
NM_003571
|
BFSP2
|
beaded filament structural protein 2, phakinin
|
|
chr7_-_27149750
|
0.486
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr9_-_138277467
|
0.486
|
NM_181701
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2
|
|
chr7_+_32501577
|
0.485
|
|
AVL9
|
AVL9 homolog (S. cerevisiase)
|
|
chr5_-_131160589
|
0.483
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr1_-_26105230
|
0.483
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr17_+_44325127
|
0.482
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr9_+_124742932
|
0.481
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr5_+_34692352
|
0.479
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr10_+_51497716
|
0.479
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr1_-_152797707
|
0.475
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr10_-_120504445
|
0.474
|
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr1_+_15725935
|
0.472
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr11_-_71491983
|
0.468
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr6_+_151815114
|
0.467
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr10_+_121642204
|
0.466
|
NM_007190
|
SEC23IP
|
SEC23 interacting protein
|
|
chr20_+_43953331
|
0.466
|
|
CTSA
|
cathepsin A
|
|
chr2_-_148494732
|
0.466
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr10_-_14090459
|
0.465
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_+_45542653
|
0.464
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr1_-_178113517
|
0.462
|
NM_022347
NM_145034
|
TOR1AIP2
|
torsin A interacting protein 2
|
|
chr10_-_120504267
|
0.459
|
|
|
|
|
chr13_-_20533703
|
0.457
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr10_+_70553993
|
0.456
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr11_+_32993973
|
0.454
|
NM_001077242
|
DEPDC7
|
DEP domain containing 7
|
|
chr5_-_131160608
|
0.454
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr3_-_135131207
|
0.452
|
NM_025041
|
C3orf36
|
chromosome 3 open reading frame 36
|
|
chr11_-_72141064
|
0.450
|
NM_001040118
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr6_-_84475424
|
0.449
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chrX_-_101284026
|
0.448
|
NM_001006938
|
TCEAL6
|
transcription elongation factor A (SII)-like 6
|
|
chr10_+_70553862
|
0.447
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr4_+_17225344
|
0.447
|
NM_025205
|
MED28
|
mediator complex subunit 28
|
|
chr3_+_185515108
|
0.445
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr15_-_38000379
|
0.445
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr5_-_133734616
|
0.444
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr8_-_126173055
|
0.438
|
|
KIAA0196
|
KIAA0196
|
|
chr20_+_30871357
|
0.438
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr7_-_95789204
|
0.436
|
NM_001160210
NM_014251
|
SLC25A13
|
solute carrier family 25, member 13 (citrin)
|
|
chr16_-_1465007
|
0.436
|
|
CLCN7
|
chloride channel 7
|
|
chr2_+_148495043
|
0.433
|
NM_018328
|
MBD5
|
methyl-CpG binding domain protein 5
|
|
chr9_+_116389814
|
0.433
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chrX_+_102356659
|
0.432
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr8_-_71743908
|
0.431
|
|
LACTB2
|
lactamase, beta 2
|
|
chr5_+_133734768
|
0.429
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr8_+_109525050
|
0.429
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr10_+_70553983
|
0.428
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_-_32054164
|
0.428
|
NM_144569
|
SPOCD1
|
SPOC domain containing 1
|
|
chr11_-_8849485
|
0.424
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_-_39859689
|
0.424
|
|
THUMPD2
|
THUMP domain containing 2
|
|
chr1_-_197173159
|
0.423
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|