|
chr21_-_26294332
|
1.083
|
|
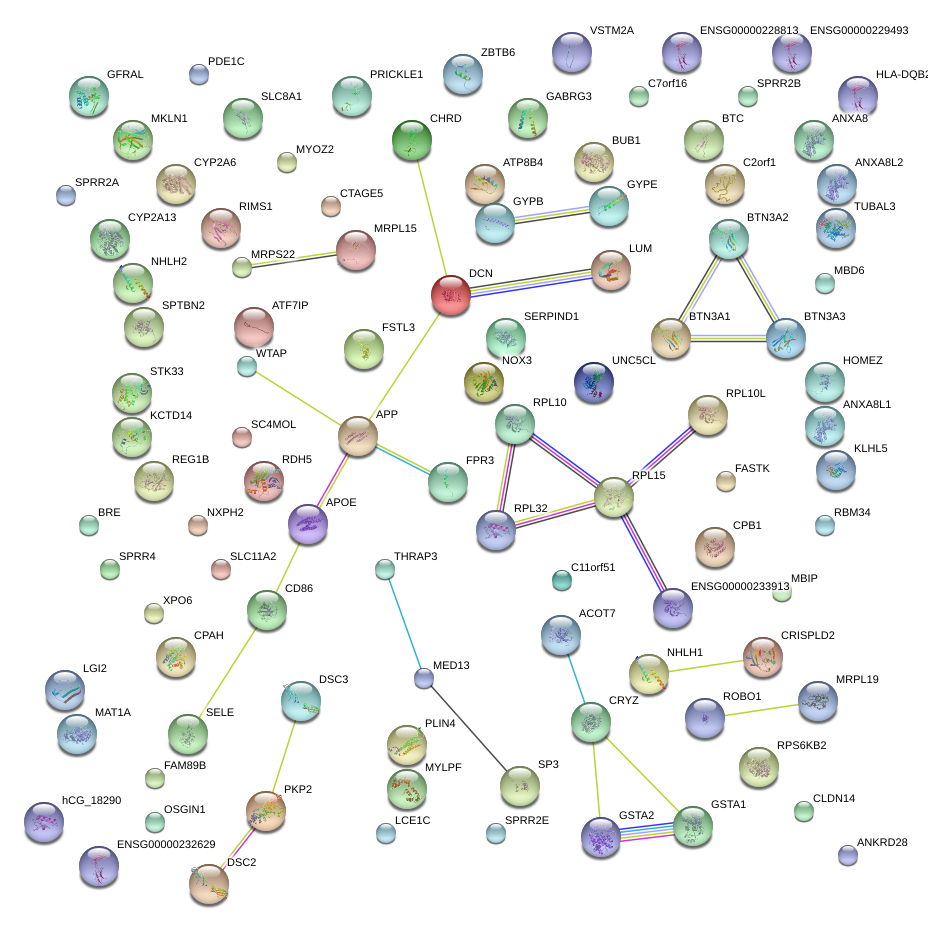
APP
|
amyloid beta (A4) precursor protein
|
|
chr1_-_151333624
|
0.946
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr6_-_32839244
|
0.934
|
NM_001198858
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr2_-_79168626
|
0.848
|
NM_006507
|
REG1B
|
regenerating islet-derived 1 beta
|
|
chr7_+_31693155
|
0.843
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr12_-_90096275
|
0.834
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr7_-_32077504
|
0.790
|
NM_001191059
NM_005020
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr2_-_174537021
|
0.729
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr21_-_36870736
|
0.708
|
NM_001146077
|
CLDN14
|
claudin 14
|
|
chr11_-_71501385
|
0.703
|
NM_014042
|
C11orf51
|
chromosome 11 open reading frame 51
|
|
chr11_-_8571988
|
0.701
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr7_+_54577512
|
0.688
|
NM_182546
|
VSTM2A
|
V-set and transmembrane domain containing 2A
|
|
chr10_-_5436781
|
0.687
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr4_+_120276386
|
0.684
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr6_+_26548678
|
0.669
|
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr6_+_131190237
|
0.659
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr10_-_46593923
|
0.658
|
NM_001040084
NM_001098845
|
ANXA8L2
LOC653110
ANXA8
ANXA8L1
|
annexin A8-like 2
hypothetical LOC653110
annexin A8
annexin A8-like 1
|
|
chr6_+_55300225
|
0.644
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr14_-_35859545
|
0.641
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr2_-_174536761
|
0.637
|
|
SP3
|
Sp3 transcription factor
|
|
chr10_-_82039158
|
0.629
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr3_-_12858006
|
0.629
|
NM_000994
NM_001007073
|
RPL32
|
ribosomal protein L32
|
|
chr6_+_73133152
|
0.622
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr6_-_41114905
|
0.608
|
NM_173561
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like
|
|
chr1_-_74971276
|
0.608
|
|
CRYZ
|
crystallin, zeta (quinone reductase)
|
|
chrX_+_153279764
|
0.579
|
NM_006013
|
RPL10
|
ribosomal protein L10
|
|
chr12_-_49708279
|
0.579
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr3_-_78749633
|
0.557
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr3_-_15814678
|
0.550
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr2_-_139254273
|
0.548
|
NM_007226
|
NXPH2
|
neurexophilin 2
|
|
chr6_-_52736231
|
0.542
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr16_-_28130662
|
0.537
|
NM_015171
|
XPO6
|
exportin 6
|
|
chr12_+_54400417
|
0.531
|
NM_002905
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis)
|
|
chr7_+_130445394
|
0.528
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr12_-_41269679
|
0.519
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr11_+_65096395
|
0.518
|
NM_001098784
NM_001098785
NM_152832
|
FAM89B
|
family with sequence similarity 89, member B
|
|
chr11_+_66952462
|
0.516
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr15_-_48198697
|
0.515
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr12_-_32940931
|
0.510
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr4_+_166468266
|
0.507
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chr1_-_167969802
|
0.504
|
NM_000450
|
SELE
|
selectin E
|
|
chr4_-_75938710
|
0.500
|
NM_001729
|
BTC
|
betacellulin
|
|
chr19_-_4468715
|
0.493
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr3_+_185580554
|
0.493
|
NM_003741
|
CHRD
|
chordin
|
|
chr12_+_56202874
|
0.491
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr7_-_150408824
|
0.486
|
NM_006712
NM_033015
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr6_+_160068135
|
0.482
|
NM_004906
NM_152858
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr4_+_38722593
|
0.480
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr7_+_37689903
|
0.479
|
|
|
|
|
chr14_-_22825072
|
0.478
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr11_-_77411933
|
0.476
|
NM_023930
|
KCTD14
|
potassium channel tetramerisation domain containing 14
|
|
chr19_+_627346
|
0.476
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr1_+_36462625
|
0.474
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr1_-_6368324
|
0.471
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr2_-_111152075
|
0.469
|
NM_004336
|
BUB1
|
budding uninhibited by benzimidazoles 1 homolog (yeast)
|
|
chr14_-_22825148
|
0.465
|
NM_020834
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr1_-_151296611
|
0.463
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr4_-_24641404
|
0.461
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr1_-_151045730
|
0.461
|
NM_178351
|
LCE1C
|
late cornified envelope 1C
|
|
chr14_+_38804226
|
0.455
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr17_-_57497380
|
0.454
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr9_-_124715392
|
0.453
|
NM_006626
|
ZBTB6
|
zinc finger and BTB domain containing 6
|
|
chr18_-_26935883
|
0.449
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr6_+_44418374
|
0.446
|
NM_145026
|
SPATS1
|
spermatogenesis associated, serine-rich 1
|
|
chr6_-_155818728
|
0.444
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr8_+_55210333
|
0.443
|
NM_014175
|
MRPL15
|
mitochondrial ribosomal protein L15
|
|
chr12_-_90029328
|
0.440
|
|
LUM
|
lumican
|
|
chr16_+_82544327
|
0.436
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr2_+_27848085
|
0.436
|
NM_004891
NM_145330
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr1_+_116181927
|
0.436
|
|
NHLH2
|
nescient helix loop helix 2
|
|
chr8_-_68821131
|
0.435
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr1_-_233391129
|
0.431
|
NM_001161533
NM_015014
|
RBM34
|
RNA binding motif protein 34
|
|
chr4_-_145281237
|
0.429
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr2_-_40593074
|
0.428
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr1_+_36462583
|
0.425
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr10_+_47216925
|
0.424
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr15_+_24799174
|
0.424
|
NM_033223
|
GABRG3
|
gamma-aminobutyric acid (GABA) A receptor, gamma 3
|
|
chr16_+_30293598
|
0.423
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr3_+_123279439
|
0.421
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr19_+_56990222
|
0.420
|
NM_002030
|
FPR3
|
formyl peptide receptor 3
|
|
chr12_+_14429499
|
0.419
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr3_+_140545528
|
0.419
|
NM_020191
|
MRPS22
|
mitochondrial ribosomal protein S22
|
|
chr16_+_83411086
|
0.418
|
NM_031476
|
CRISPLD2
|
cysteine-rich secretory protein LCCL domain containing 2
|
|
chr11_-_66245288
|
0.417
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr6_+_26473376
|
0.415
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr1_+_151209751
|
0.412
|
NM_173080
|
SPRR4
|
small proline-rich protein 4
|
|
chr5_-_13997588
|
0.402
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr2_-_233060774
|
0.401
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr8_-_18710575
|
0.399
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_-_114925734
|
0.398
|
NM_005872
|
BCAS2
|
breast carcinoma amplified sequence 2
|
|
chr5_+_146594718
|
0.395
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr16_+_88224175
|
0.393
|
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr17_-_78391178
|
0.390
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr1_-_94475504
|
0.390
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr15_-_24569201
|
0.389
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr1_+_144149794
|
0.389
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr3_-_192063158
|
0.386
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr17_+_40494201
|
0.386
|
NM_021079
|
NMT1
|
N-myristoyltransferase 1
|
|
chr6_-_33822659
|
0.386
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr15_-_32289561
|
0.384
|
NM_024713
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr3_+_101462375
|
0.381
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr8_-_91726915
|
0.381
|
|
TMEM64
|
transmembrane protein 64
|
|
chr20_-_61356247
|
0.380
|
NM_152864
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4
|
|
chr13_+_113023938
|
0.378
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr15_+_46285789
|
0.378
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr22_-_35914200
|
0.377
|
NM_031910
NM_182486
|
C1QTNF6
|
C1q and tumor necrosis factor related protein 6
|
|
chr1_+_148020869
|
0.376
|
NM_000566
|
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64)
|
|
chr20_+_2224612
|
0.376
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr1_-_202449659
|
0.375
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr21_-_38954433
|
0.375
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr8_+_133948384
|
0.375
|
NM_003235
|
TG
|
thyroglobulin
|
|
chr11_-_127962662
|
0.371
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr5_-_40871082
|
0.370
|
NM_000997
|
RPL37
|
ribosomal protein L37
|
|
chr2_-_217433018
|
0.370
|
NM_003284
|
TNP1
|
transition protein 1 (during histone to protamine replacement)
|
|
chr6_-_24597717
|
0.370
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chrX_+_154097894
|
0.369
|
NM_003372
|
VBP1
|
von Hippel-Lindau binding protein 1
|
|
chr7_-_26198673
|
0.368
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr12_+_32578513
|
0.366
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr10_+_76541351
|
0.366
|
NM_001174156
NM_144660
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr2_+_207215386
|
0.363
|
NM_001102659
|
LOC200726
|
hCG1657980
|
|
chr7_+_142692165
|
0.361
|
NM_153345
|
TMEM139
|
transmembrane protein 139
|
|
chr17_+_38412267
|
0.360
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr7_+_100156358
|
0.359
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr4_-_38534802
|
0.359
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr7_+_133982070
|
0.359
|
NM_001724
NM_199186
|
BPGM
|
2,3-bisphosphoglycerate mutase
|
|
chr1_+_184532027
|
0.358
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr2_-_200038055
|
0.357
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr1_+_23890880
|
0.357
|
NM_000975
|
RPL11
|
ribosomal protein L11
|
|
chr2_-_55413253
|
0.356
|
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr5_+_173405343
|
0.355
|
|
HMP19
|
HMP19 protein
|
|
chr6_-_66473790
|
0.354
|
NM_001142800
NM_001142801
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr18_+_27332024
|
0.351
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr9_-_115877104
|
0.350
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr17_-_46140015
|
0.350
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr21_+_17841297
|
0.349
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr12_-_6450015
|
0.348
|
NM_014231
NM_016830
NM_199245
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1)
|
|
chr2_-_220144254
|
0.348
|
|
OBSL1
|
obscurin-like 1
|
|
chr18_+_27281729
|
0.346
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr4_-_73653331
|
0.345
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr4_+_86968069
|
0.345
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr12_-_48508411
|
0.344
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chrX_+_109132507
|
0.344
|
NM_017698
|
TMEM164
|
transmembrane protein 164
|
|
chr18_+_54013575
|
0.343
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr8_-_97316933
|
0.342
|
NM_006294
|
UQCRB
|
ubiquinol-cytochrome c reductase binding protein
|
|
chr2_+_27848090
|
0.341
|
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr10_-_116408046
|
0.340
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr15_-_32289480
|
0.340
|
|
C15orf29
|
chromosome 15 open reading frame 29
|
|
chr6_+_31059463
|
0.339
|
NM_001010909
|
MUC21
|
mucin 21, cell surface associated
|
|
chr17_+_38403935
|
0.339
|
NM_000988
|
RPL27
|
ribosomal protein L27
|
|
chr1_-_94475840
|
0.338
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr10_+_86174665
|
0.338
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr1_-_24342158
|
0.337
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr6_-_35804337
|
0.337
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr10_-_74863303
|
0.337
|
NM_001024593
|
ZMYND17
|
zinc finger, MYND-type containing 17
|
|
chr16_+_70437394
|
0.336
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chrX_-_30817329
|
0.334
|
NM_152787
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr10_+_123744131
|
0.334
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chrX_-_31194867
|
0.333
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr6_-_32265489
|
0.332
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr1_+_48460943
|
0.332
|
NM_001011547
NM_001135181
|
SLC5A9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9
|
|
chr1_-_239869826
|
0.331
|
NM_014322
|
OPN3
|
opsin 3
|
|
chr9_-_32563057
|
0.330
|
NM_002493
NM_182739
|
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa
|
|
chr15_-_24569253
|
0.327
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr7_-_150408785
|
0.327
|
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr8_+_38877909
|
0.326
|
NM_021623
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr8_+_32905027
|
0.325
|
|
|
|
|
chr1_+_195213289
|
0.323
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr11_+_63766163
|
0.323
|
NM_057092
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr7_+_93373755
|
0.322
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr21_+_40161216
|
0.322
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chr19_+_45564549
|
0.321
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr6_+_112515366
|
0.320
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr1_+_23890857
|
0.318
|
|
RPL11
|
ribosomal protein L11
|
|
chr1_+_1360765
|
0.318
|
NM_022834
NM_199121
|
VWA1
|
von Willebrand factor A domain containing 1
|
|
chr1_+_112811562
|
0.318
|
NM_004185
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr15_-_24569309
|
0.317
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr3_-_122951200
|
0.317
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr3_-_180272229
|
0.316
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr5_+_173405298
|
0.316
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr17_+_58440629
|
0.315
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr3_-_168935287
|
0.314
|
NM_007217
NM_145860
NM_145859
|
PDCD10
|
programmed cell death 10
|
|
chr1_+_116744992
|
0.313
|
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr3_+_153469421
|
0.312
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr21_-_18697829
|
0.311
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr14_+_23008723
|
0.310
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr3_+_2115449
|
0.310
|
|
CNTN4
|
contactin 4
|
|
chr1_-_203985963
|
0.309
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr16_-_28130201
|
0.308
|
|
XPO6
|
exportin 6
|
|
chr2_+_201099093
|
0.308
|
NM_001160033
NM_001160046
NM_152524
|
SGOL2
|
shugoshin-like 2 (S. pombe)
|
|
chr1_-_155788775
|
0.308
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr4_+_71418886
|
0.307
|
NM_212557
|
AMTN
|
amelotin
|
|
chr19_+_40474867
|
0.307
|
NM_002361
NM_080600
|
MAG
|
myelin associated glycoprotein
|
|
chr10_-_71753684
|
0.307
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr6_-_99979850
|
0.306
|
NM_015491
NM_032870
|
SFRS18
|
splicing factor, arginine/serine-rich 18
|
|
chr20_+_42537927
|
0.306
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr20_+_56318415
|
0.306
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chrX_-_131375217
|
0.306
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr2_+_113119985
|
0.302
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr10_-_71713455
|
0.301
|
NM_022146
|
NPFFR1
|
neuropeptide FF receptor 1
|
|
chr6_+_31741814
|
0.299
|
|
CSNK2B
|
casein kinase 2, beta polypeptide
|