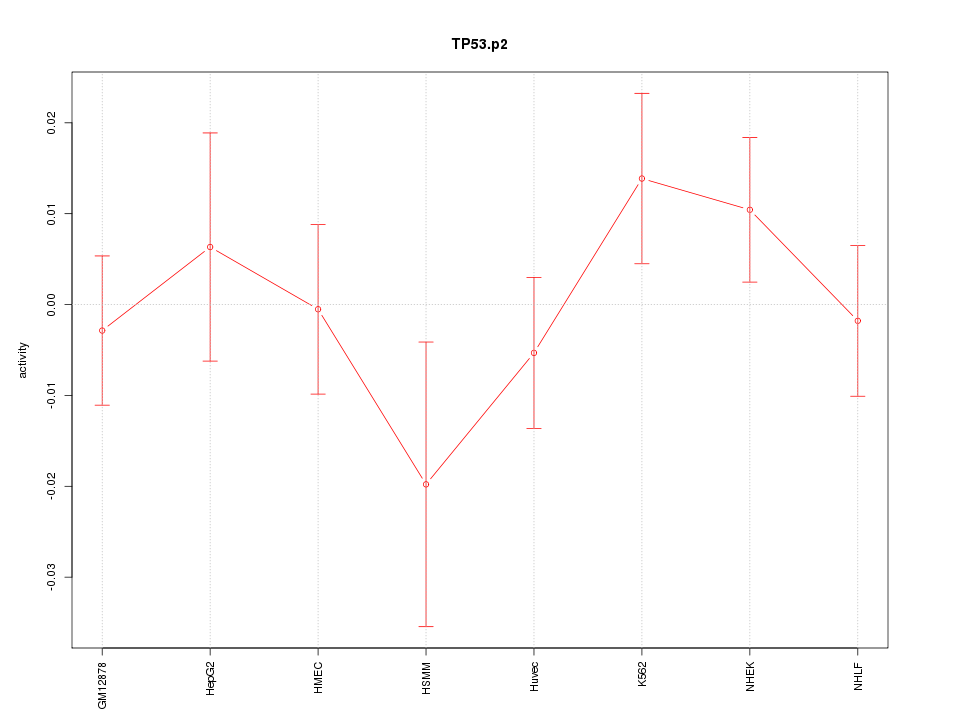
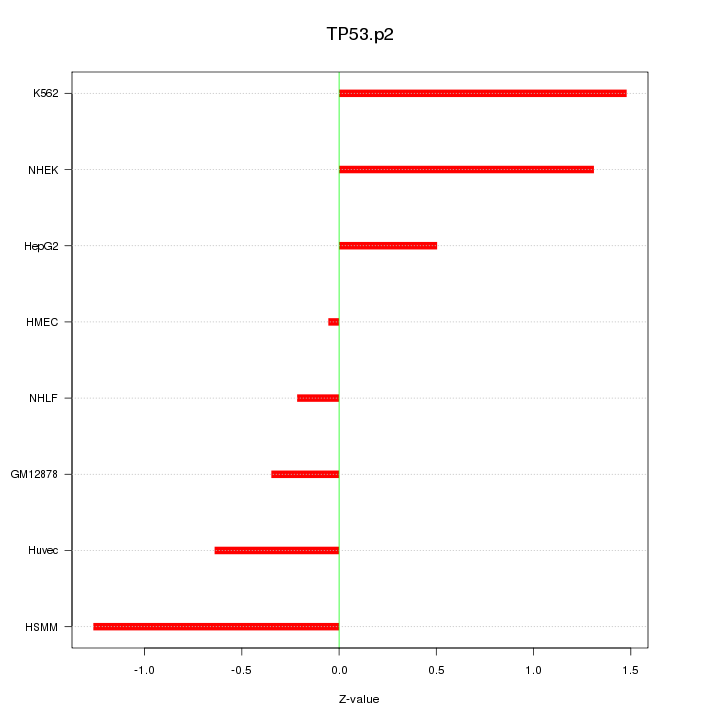
|
chr6_+_36752244
|
1.367
|
NM_078467
|
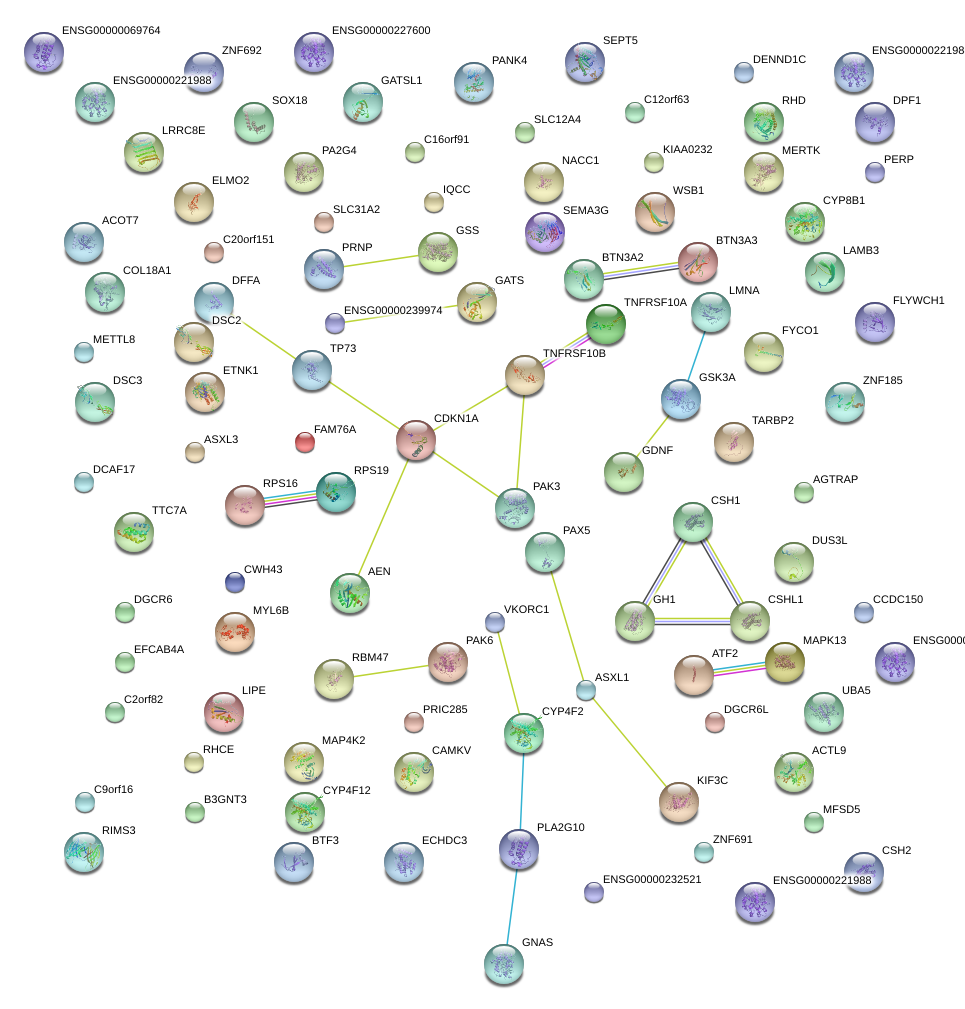
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr19_-_8670054
|
1.036
|
NM_178525
|
ACTL9
|
actin-like 9
|
|
chr1_+_32443822
|
0.989
|
NM_001160042
NM_018134
|
IQCC
|
IQ motif containing C
|
|
chr19_-_47619992
|
0.953
|
|
LIPE
|
lipase, hormone-sensitive
|
|
chr12_+_54832470
|
0.879
|
NM_002475
|
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle
|
|
chr22_-_18687511
|
0.765
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr7_+_72206961
|
0.759
|
|
GTF2IP1
|
general transcription factor IIi, pseudogene 1
|
|
chr8_-_23138517
|
0.728
|
|
TNFRSF10A
|
tumor necrosis factor receptor superfamily, member 10a
|
|
chr6_-_138470160
|
0.697
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr9_+_114953087
|
0.683
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr10_+_11824361
|
0.660
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr1_+_3597092
|
0.653
|
NM_001126240
NM_001126241
NM_001126242
|
TP73
|
tumor protein p73
|
|
chr2_+_171998988
|
0.648
|
NM_001164821
NM_025000
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr2_-_171998681
|
0.629
|
|
METTL8
|
methyltransferase like 8
|
|
chr18_-_26876622
|
0.603
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr16_-_1410710
|
0.596
|
|
C16orf91
|
chromosome 16 open reading frame 91
|
|
chr16_-_31013626
|
0.590
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr2_+_197212522
|
0.582
|
NM_001080539
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr2_-_26058941
|
0.566
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr20_-_62151216
|
0.559
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr2_-_120697382
|
0.546
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr4_-_40211324
|
0.543
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr16_-_14695930
|
0.538
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr1_-_10455100
|
0.534
|
|
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide
|
|
chr4_+_48683021
|
0.523
|
NM_025087
|
CWH43
|
cell wall biogenesis 43 C-terminal homolog (S. cerevisiae)
|
|
chr2_+_171999042
|
0.516
|
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr19_-_47438583
|
0.509
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr19_-_43406621
|
0.498
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr20_-_33007243
|
0.478
|
NM_000178
|
GSS
|
glutathione synthetase
|
|
chr1_-_10455190
|
0.471
|
NM_004401
NM_213566
|
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide
|
|
chr12_+_22669352
|
0.464
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr1_-_207892420
|
0.459
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr2_-_26058831
|
0.457
|
|
KIF3C
|
kinesin family member 3C
|
|
chr2_+_47021816
|
0.454
|
NM_020458
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr2_-_175741096
|
0.446
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr1_+_43084846
|
0.441
|
NM_015911
|
ZNF691
|
zinc finger protein 691
|
|
chr9_+_129962292
|
0.431
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr15_+_86965526
|
0.421
|
NM_022767
|
AEN
|
apoptosis enhancing nuclease
|
|
chr20_-_33007242
|
0.402
|
|
GSS
|
glutathione synthetase
|
|
chr17_-_59304729
|
0.401
|
NM_020991
NM_022644
NM_022645
|
CSH2
|
chorionic somatomammotropin hormone 2
|
|
chr7_-_74291367
|
0.396
|
|
GTF2IP1
|
general transcription factor IIi, pseudogene 1
|
|
chr19_+_15644885
|
0.394
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr20_-_33007209
|
0.390
|
|
GSS
|
glutathione synthetase
|
|
chr15_+_38319326
|
0.390
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr1_-_40903897
|
0.386
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr20_-_60435983
|
0.384
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr8_-_22982351
|
0.381
|
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr19_-_47438518
|
0.371
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_+_47021866
|
0.367
|
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr19_-_47438563
|
0.366
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr16_-_66555449
|
0.363
|
NM_001145963
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr19_+_47056162
|
0.357
|
|
RPS19
|
ribosomal protein S19
|
|
chr19_+_17766907
|
0.357
|
NM_014256
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3
|
|
chr2_+_233443237
|
0.354
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr1_-_6375949
|
0.350
|
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr20_+_56900116
|
0.349
|
NM_001077488
|
GNAS
|
GNAS complex locus
|
|
chr20_-_61676003
|
0.345
|
NM_001037335
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr19_-_15869870
|
0.340
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr19_-_47438548
|
0.337
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr1_+_11718764
|
0.336
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr20_-_44468607
|
0.335
|
|
ELMO2
|
engulfment and cell motility 2
|
|
chr4_+_6835388
|
0.334
|
|
KIAA0232
|
KIAA0232
|
|
chr7_-_74705276
|
0.330
|
NM_001145064
|
GATSL2
|
GATS protein-like 2
|
|
chr12_+_54784681
|
0.324
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr19_+_7859380
|
0.323
|
NM_025061
|
LRRC8E
|
leucine rich repeat containing 8 family, member E
|
|
chr12_+_51932136
|
0.316
|
NM_032889
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr1_+_154318954
|
0.314
|
|
LMNA
|
lamin A/C
|
|
chr19_+_47056102
|
0.312
|
|
RPS19
|
ribosomal protein S19
|
|
chr12_+_52181643
|
0.311
|
NM_134324
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr1_+_27925054
|
0.305
|
NM_001143912
NM_001143913
NM_001143914
NM_001143915
NM_152660
|
FAM76A
|
family with sequence similarity 76, member A
|
|
chr19_+_17766935
|
0.304
|
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3
|
|
chr12_+_51932252
|
0.303
|
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr20_-_44468641
|
0.303
|
|
ELMO2
|
engulfment and cell motility 2
|
|
chr16_+_2901995
|
0.301
|
|
FLYWCH1
|
FLYWCH-type zinc finger 1
|
|
chr21_+_45649429
|
0.298
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr19_+_13090101
|
0.296
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr1_-_2447848
|
0.293
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr2_+_171999082
|
0.292
|
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr4_+_6726699
|
0.291
|
|
LOC93622
|
hypothetical LOC93622
|
|
chr18_+_29412538
|
0.288
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr1_+_25471470
|
0.282
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr1_+_11718798
|
0.281
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr11_-_64327197
|
0.277
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr3_-_42892195
|
0.276
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr19_-_44618375
|
0.276
|
NM_001020
|
RPS16
|
ribosomal protein S16
|
|
chr6_+_32229599
|
0.275
|
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr17_+_22645268
|
0.273
|
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr3_-_52454082
|
0.273
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr19_-_6432765
|
0.271
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr19_-_5741913
|
0.270
|
|
DUS3L
|
dihydrouridine synthase 3-like (S. cerevisiae)
|
|
chr22_+_18085990
|
0.268
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr20_+_30409649
|
0.267
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr2_+_112372651
|
0.267
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr1_-_247119516
|
0.266
|
NM_001136036
|
ZNF692
|
zinc finger protein 692
|
|
chr4_-_9727384
|
0.264
|
|
|
|
|
chr11_+_817584
|
0.264
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr8_-_22982435
|
0.262
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr3_-_46012303
|
0.262
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr6_+_36206239
|
0.261
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chrX_+_151837064
|
0.261
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr4_+_153676857
|
0.260
|
|
DKFZP434I0714
|
hypothetical protein DKFZP434I0714
|
|
chr5_+_72830029
|
0.258
|
|
BTF3
|
basic transcription factor 3
|
|
chr5_-_131854325
|
0.251
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr1_+_11718715
|
0.251
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr8_-_145671947
|
0.249
|
|
FOXH1
|
forkhead box H1
|
|
chr12_+_51932173
|
0.248
|
|
MFSD5
|
major facilitator superfamily domain containing 5
|
|
chr12_-_46385991
|
0.247
|
|
RPAP3
|
RNA polymerase II associated protein 3
|
|
chr22_+_21852551
|
0.244
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr4_+_6835363
|
0.242
|
|
KIAA0232
|
KIAA0232
|
|
chr22_-_16637221
|
0.241
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr15_-_62242145
|
0.240
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr12_+_54784632
|
0.239
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr12_-_14612057
|
0.238
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr1_-_54291634
|
0.235
|
NM_004872
|
TMEM59
|
transmembrane protein 59
|
|
chr5_-_175721320
|
0.234
|
NM_001079684
NM_001079685
NM_020444
|
KIAA1191
|
KIAA1191
|
|
chr12_+_6808025
|
0.231
|
|
LEPREL2
|
leprecan-like 2
|
|
chr20_-_23755297
|
0.231
|
NM_001322
|
CST2
|
cystatin SA
|
|
chr1_+_23568028
|
0.229
|
|
C1orf213
|
chromosome 1 open reading frame 213
|
|
chr22_-_16637160
|
0.229
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr19_+_43557264
|
0.229
|
|
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8
|
|
chr16_+_387176
|
0.227
|
NM_005009
|
NME4
|
non-metastatic cells 4, protein expressed in
|
|
chr9_-_168913
|
0.226
|
NM_001145355
NM_001145356
NM_018491
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr20_+_61760084
|
0.222
|
NM_016434
NM_032957
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chr1_+_45578464
|
0.220
|
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr3_+_14141633
|
0.219
|
|
TMEM43
|
transmembrane protein 43
|
|
chr15_-_62242259
|
0.219
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr2_+_169021039
|
0.218
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr15_-_62242243
|
0.218
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr19_+_54149941
|
0.217
|
|
BAX
|
BCL2-associated X protein
|
|
chr8_-_145672525
|
0.215
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chr1_-_54291474
|
0.213
|
|
TMEM59
|
transmembrane protein 59
|
|
chr6_+_33486750
|
0.213
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr7_+_100792841
|
0.213
|
NM_133457
|
EMID2
|
EMI domain containing 2
|
|
chr19_+_54149963
|
0.213
|
|
BAX
|
BCL2-associated X protein
|
|
chr1_-_114249158
|
0.212
|
NM_006594
|
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit
|
|
chr22_+_23753940
|
0.212
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr10_-_118755077
|
0.212
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr7_-_72077805
|
0.211
|
NM_198853
|
TRIM73
TRIM74
|
tripartite motif containing 73
tripartite motif containing 74
|
|
chr3_+_161601040
|
0.209
|
NM_005496
|
SMC4
|
structural maintenance of chromosomes 4
|
|
chr4_+_6835354
|
0.208
|
NM_001100590
NM_014743
|
KIAA0232
|
KIAA0232
|
|
chr1_-_54291378
|
0.207
|
|
TMEM59
|
transmembrane protein 59
|
|
chr15_-_40173949
|
0.207
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr15_-_62242237
|
0.207
|
|
PPIB
|
peptidylprolyl isomerase B (cyclophilin B)
|
|
chr3_+_14141583
|
0.206
|
|
TMEM43
|
transmembrane protein 43
|
|
chr22_-_22422951
|
0.206
|
NM_021916
|
ZNF70
|
zinc finger protein 70
|
|
chr16_-_31013594
|
0.206
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr11_-_9243408
|
0.206
|
NM_015213
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr22_-_16637194
|
0.205
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr12_+_97563049
|
0.205
|
NM_001160
NM_013229
NM_181861
NM_181868
NM_181869
|
APAF1
|
apoptotic peptidase activating factor 1
|
|
chr1_-_25619863
|
0.199
|
NM_020485
NM_138616
NM_138617
NM_138618
|
RHCE
RHD
|
Rh blood group, CcEe antigens
Rh blood group, D antigen
|
|
chr22_-_21231421
|
0.197
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr1_-_45578193
|
0.197
|
NM_001048174
NM_001048172
NM_001048173
NM_001048171
NM_001128425
NM_012222
|
MUTYH
|
mutY homolog (E. coli)
|
|
chr4_-_9727394
|
0.196
|
|
WDR1
|
WD repeat domain 1
|
|
chr20_+_29931257
|
0.194
|
|
TTLL9
|
tubulin tyrosine ligase-like family, member 9
|
|
chr19_-_18515376
|
0.194
|
NM_012181
|
FKBP8
|
FK506 binding protein 8, 38kDa
|
|
chr7_-_72380020
|
0.193
|
NM_178125
|
TRIM50
|
tripartite motif containing 50
|
|
chr11_-_107969554
|
0.191
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr8_-_10625352
|
0.191
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr3_+_9766901
|
0.190
|
|
OGG1
|
8-oxoguanine DNA glycosylase
|
|
chr8_+_23016271
|
0.190
|
NM_003841
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain
|
|
chr2_+_169021001
|
0.188
|
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr19_+_17766609
|
0.188
|
|
B3GNT3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3
|
|
chr16_-_15057328
|
0.187
|
NM_173474
|
NTAN1
|
N-terminal asparagine amidase
|
|
chr1_-_247119908
|
0.187
|
NM_001193328
NM_017865
|
ZNF692
|
zinc finger protein 692
|
|
chr1_-_54291503
|
0.186
|
|
TMEM59
|
transmembrane protein 59
|
|
chr21_+_29593607
|
0.186
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr8_-_57286412
|
0.185
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr3_-_186454509
|
0.185
|
NM_001166415
NM_001966
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase
|
|
chr3_-_24511456
|
0.184
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr11_+_63758550
|
0.183
|
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr1_+_226261374
|
0.182
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr11_+_93917214
|
0.182
|
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr20_+_56899904
|
0.181
|
|
GNAS
|
GNAS complex locus
|
|
chr17_+_69939130
|
0.181
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr19_+_778821
|
0.181
|
NM_001700
|
AZU1
|
azurocidin 1
|
|
chr12_+_54784614
|
0.180
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr12_+_54784625
|
0.179
|
|
PA2G4
|
proliferation-associated 2G4, 38kDa
|
|
chr20_+_34667566
|
0.179
|
NM_018840
NM_199483
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr4_-_9727456
|
0.177
|
|
WDR1
|
WD repeat domain 1
|
|
chr6_+_117910472
|
0.176
|
NM_173674
|
DCBLD1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr18_-_32662942
|
0.176
|
|
C18orf10
|
chromosome 18 open reading frame 10
|
|
chr22_-_29231597
|
0.175
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr4_+_6726555
|
0.175
|
|
LOC93622
|
hypothetical LOC93622
|
|
chr19_+_32976717
|
0.175
|
|
|
|
|
chr1_+_45578370
|
0.174
|
|
TOE1
|
target of EGR1, member 1 (nuclear)
|
|
chr3_+_14141553
|
0.173
|
|
TMEM43
|
transmembrane protein 43
|
|
chr22_+_27498679
|
0.173
|
|
CCDC117
|
coiled-coil domain containing 117
|
|
chr7_+_72380090
|
0.172
|
NM_001135211
NM_003602
|
FKBP6
|
FK506 binding protein 6, 36kDa
|
|
chr4_-_9727598
|
0.171
|
NM_005112
NM_017491
|
WDR1
|
WD repeat domain 1
|
|
chr11_-_47356562
|
0.170
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr19_-_32976642
|
0.170
|
|
LOC148189
|
hypothetical LOC148189
|
|
chr11_-_47356479
|
0.169
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr9_-_93225961
|
0.168
|
NM_005384
|
NFIL3
|
nuclear factor, interleukin 3 regulated
|
|
chr3_+_14141544
|
0.167
|
|
TMEM43
|
transmembrane protein 43
|
|
chr9_-_114814286
|
0.166
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr4_+_52403922
|
0.166
|
NM_001040402
NM_015115
|
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr22_-_16637238
|
0.165
|
NM_001196
NM_197967
|
BID
|
BH3 interacting domain death agonist
|
|
chr16_+_88280594
|
0.165
|
|
CDK10
|
cyclin-dependent kinase 10
|
|
chr18_+_32663056
|
0.164
|
NM_020776
|
KIAA1328
|
KIAA1328
|
|
chr5_-_10360873
|
0.164
|
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|