|
chr8_-_145097031
|
2.505
|
NM_201380
|
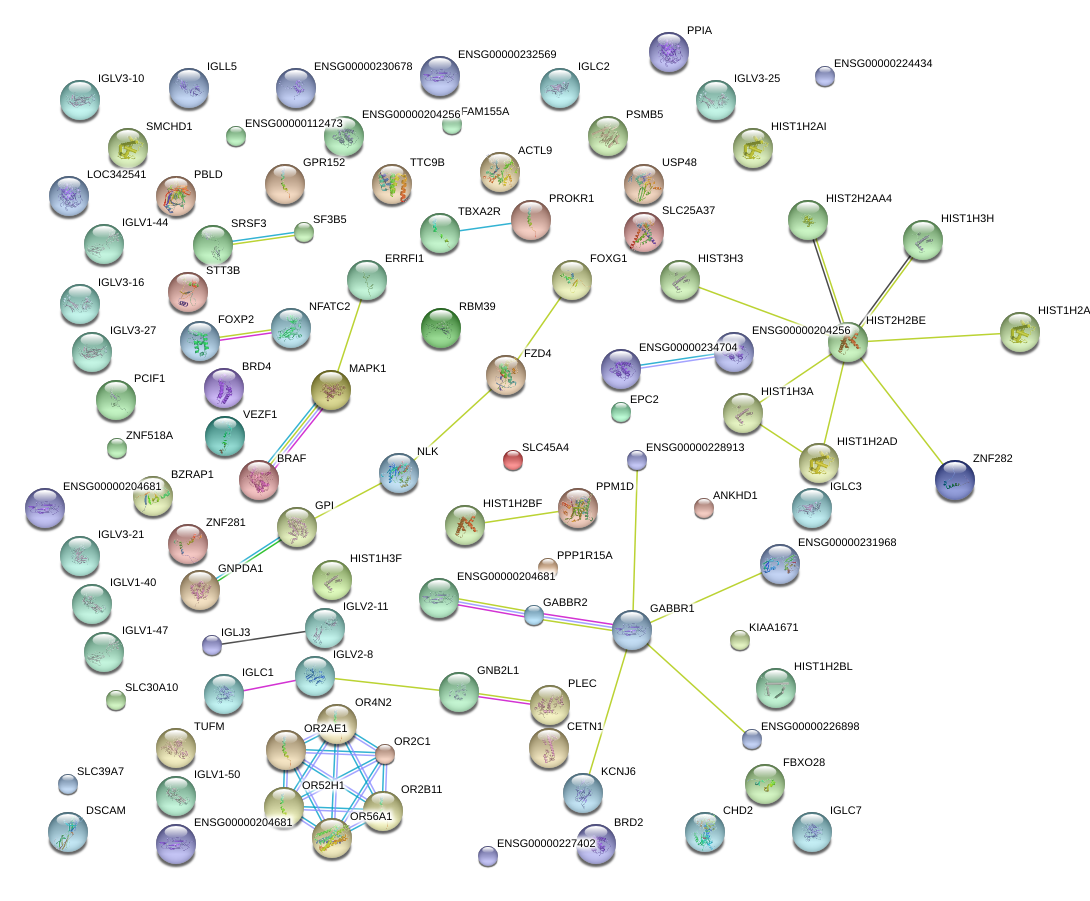
PLEC
|
plectin
|
|
chr2_+_68726457
|
2.267
|
NM_138964
|
PROKR1
|
prokineticin receptor 1
|
|
chr16_+_3345889
|
1.684
|
NM_012368
|
OR2C1
|
olfactory receptor, family 2, subfamily C, member 1
|
|
chr6_+_33047952
|
1.682
|
|
BRD2
|
bromodomain containing 2
|
|
chr14_-_22574193
|
1.663
|
NM_001144932
NM_002797
NM_001130725
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr22_+_21359175
|
1.624
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr7_-_99312591
|
1.530
|
NM_001005276
|
OR2AE1
|
olfactory receptor, family 2, subfamily AE, member 1
|
|
chr6_+_33276580
|
1.428
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr22_+_21393107
|
1.365
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr7_-_140270749
|
1.346
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr18_+_570356
|
1.335
|
NM_004066
|
CETN1
|
centrin, EF-hand protein, 1
|
|
chr22_+_21094094
|
1.309
|
|
|
|
|
chr11_-_66976775
|
1.300
|
NM_206997
|
GPR152
|
G protein-coupled receptor 152
|
|
chr2_+_149118791
|
1.287
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_+_149119029
|
1.223
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr22_+_21079355
|
1.214
|
|
|
|
|
chr6_+_36670113
|
1.163
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr22_+_23753940
|
1.161
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr14_+_19365447
|
1.127
|
NM_001004723
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2
|
|
chr17_+_56032385
|
1.091
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr8_-_142333339
|
1.090
|
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr6_+_36670067
|
1.086
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr6_+_36670117
|
1.058
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr22_+_21042091
|
1.052
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr22_-_20551722
|
1.019
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr5_-_180603335
|
1.007
|
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr7_+_148523486
|
1.007
|
NM_003575
|
ZNF282
|
zinc finger protein 282
|
|
chr19_-_3551576
|
1.001
|
|
TBXA2R
|
thromboxane A2 receptor
|
|
chr3_+_31549123
|
0.999
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr21_-_38210531
|
0.990
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr6_-_27883662
|
0.975
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr17_+_53761293
|
0.961
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr20_-_33793507
|
0.945
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr22_-_20551953
|
0.941
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr5_+_139856688
|
0.940
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chr14_-_22573764
|
0.915
|
|
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5
|
|
chr19_-_8670054
|
0.914
|
NM_178525
|
ACTL9
|
actin-like 9
|
|
chr1_-_245681906
|
0.908
|
NM_001004492
|
OR2B11
|
olfactory receptor, family 2, subfamily B, member 11
|
|
chr5_-_180603476
|
0.887
|
NM_006098
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr17_-_53420595
|
0.885
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr20_+_43996717
|
0.858
|
NM_022104
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1
|
|
chr6_+_27883877
|
0.857
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr22_+_21116283
|
0.850
|
|
|
|
|
chr17_+_23393744
|
0.847
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr9_-_100510657
|
0.843
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr14_+_28306633
|
0.842
|
|
FOXG1
|
forkhead box G1
|
|
chr6_-_29703725
|
0.833
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr10_+_97879645
|
0.830
|
|
ZNF518A
|
zinc finger protein 518A
|
|
chr22_+_21065134
|
0.830
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr11_-_5523328
|
0.825
|
NM_001005289
|
OR52H1
|
olfactory receptor, family 52, subfamily H, member 1
|
|
chr22_+_21065202
|
0.821
|
|
|
|
|
chr15_+_91244563
|
0.811
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr7_+_44802834
|
0.809
|
|
PPIA
|
peptidylprolyl isomerase A (cyclophilin A)
|
|
chr19_-_45416112
|
0.783
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr15_+_91244422
|
0.776
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr10_-_69762654
|
0.775
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr22_+_21065207
|
0.775
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr20_-_49612574
|
0.771
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr11_-_6005546
|
0.766
|
NM_001001917
|
OR56A1
|
olfactory receptor, family 56, subfamily A, member 1
|
|
chr1_-_218168404
|
0.763
|
|
|
|
|
chr1_-_21982037
|
0.741
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr7_+_113842284
|
0.727
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr1_-_226679648
|
0.717
|
NM_003493
|
HIST3H3
|
histone cluster 3, H3
|
|
chr7_+_44802798
|
0.716
|
|
PPIA
|
peptidylprolyl isomerase A (cyclophilin A)
|
|
chr11_-_86343859
|
0.713
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr17_-_53761119
|
0.712
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr8_+_23442413
|
0.712
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr16_-_28765138
|
0.707
|
NM_003321
|
TUFM
|
Tu translation elongation factor, mitochondrial
|
|
chr1_+_222368413
|
0.704
|
NM_001136115
NM_015176
|
FBXO28
|
F-box protein 28
|
|
chr19_+_54068373
|
0.702
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr6_-_144458446
|
0.701
|
NM_031287
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr13_-_107317460
|
0.699
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr1_-_218168137
|
0.695
|
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr1_-_218168548
|
0.693
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr1_-_7998265
|
0.685
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr1_-_198644111
|
0.683
|
|
ZNF281
|
zinc finger protein 281
|
|
chr1_-_159369041
|
0.681
|
|
DEDD
|
death effector domain containing
|
|
chr9_-_129869211
|
0.679
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr2_+_127892459
|
0.677
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr14_+_19681734
|
0.673
|
NM_001004724
|
OR4N5
|
olfactory receptor, family 4, subfamily N, member 5
|
|
chr19_+_10673105
|
0.670
|
NM_031209
|
QTRT1
|
queuine tRNA-ribosyltransferase 1
|
|
chr19_+_54068300
|
0.666
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr2_+_48521411
|
0.663
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr8_+_23442331
|
0.660
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr8_+_23442307
|
0.656
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr7_+_154493478
|
0.655
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr1_+_114323552
|
0.653
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr6_-_74284522
|
0.647
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr20_-_49612745
|
0.645
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr20_-_33793286
|
0.645
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr6_-_37333872
|
0.640
|
NM_001162900
|
TMEM217
|
transmembrane protein 217
|
|
chr17_-_39630951
|
0.639
|
NM_001098833
NM_020218
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr9_+_130124873
|
0.638
|
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chr3_+_31549435
|
0.637
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr17_-_24645202
|
0.633
|
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr22_+_21053972
|
0.631
|
|
|
|
|
chr14_+_59934178
|
0.631
|
|
|
|
|
chr6_-_33276416
|
0.630
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr6_-_31617748
|
0.626
|
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B
|
|
chr7_+_104441730
|
0.624
|
NM_018682
NM_182931
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr1_-_159369081
|
0.624
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr6_+_105511615
|
0.622
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr16_-_30274182
|
0.622
|
NM_006110
|
CD2BP2
|
CD2 (cytoplasmic tail) binding protein 2
|
|
chr10_+_97879457
|
0.621
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr8_-_144762765
|
0.620
|
NM_023078
|
PYCRL
|
pyrroline-5-carboxylate reductase-like
|
|
chr17_+_72244224
|
0.615
|
|
MFSD11
|
major facilitator superfamily domain containing 11
|
|
chr9_-_130124820
|
0.614
|
|
TRUB2
|
TruB pseudouridine (psi) synthase homolog 2 (E. coli)
|
|
chr2_+_70168487
|
0.614
|
|
|
|
|
chr12_+_54361685
|
0.613
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr1_+_51208229
|
0.609
|
NM_078626
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr14_+_102636233
|
0.609
|
NM_001077594
|
EXOC3L4
|
exocyst complex component 3-like 4
|
|
chr2_-_236741352
|
0.605
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr17_-_3283884
|
0.605
|
NM_003554
|
OR1E2
|
olfactory receptor, family 1, subfamily E, member 2
|
|
chr2_+_46623396
|
0.601
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr1_-_152431175
|
0.598
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr3_-_12680612
|
0.598
|
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr3_+_31549478
|
0.598
|
NM_178862
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr22_+_19601940
|
0.597
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr1_-_159368985
|
0.597
|
|
DEDD
|
death effector domain containing
|
|
chr1_+_51208499
|
0.596
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr6_-_33276426
|
0.595
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr2_+_236068029
|
0.594
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr19_-_15390816
|
0.592
|
NM_014371
|
AKAP8L
|
A kinase (PRKA) anchor protein 8-like
|
|
chr7_+_128651976
|
0.591
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr6_-_26324850
|
0.591
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr7_+_128099581
|
0.589
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr6_-_33276347
|
0.586
|
NM_021976
|
RXRB
|
retinoid X receptor, beta
|
|
chr19_-_12858956
|
0.584
|
NM_006563
|
KLF1
|
Kruppel-like factor 1 (erythroid)
|
|
chr22_+_21011653
|
0.583
|
|
|
|
|
chr15_+_48261649
|
0.582
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr1_+_166172420
|
0.582
|
NM_001017977
NM_001198956
NM_001198957
NM_018442
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr22_-_17799177
|
0.580
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr6_-_2696152
|
0.580
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr6_+_30799130
|
0.579
|
|
TUBB
|
tubulin, beta
|
|
chr9_+_35595275
|
0.576
|
NM_006285
|
TESK1
|
testis-specific kinase 1
|
|
chr19_-_15390738
|
0.574
|
|
AKAP8L
|
A kinase (PRKA) anchor protein 8-like
|
|
chr6_-_39305203
|
0.572
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr2_+_170298700
|
0.571
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr6_+_31891509
|
0.570
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr19_-_45416043
|
0.567
|
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr22_+_21370394
|
0.566
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr12_+_54361596
|
0.562
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr4_-_1980756
|
0.561
|
NM_005663
|
WHSC2
|
Wolf-Hirschhorn syndrome candidate 2
|
|
chr8_-_107023
|
0.560
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr12_-_70343790
|
0.560
|
|
ZFC3H1
|
zinc finger, C3H1-type containing
|
|
chr22_+_21340757
|
0.559
|
|
|
|
|
chr22_-_17799215
|
0.559
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr5_+_140454382
|
0.556
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr6_-_31617704
|
0.554
|
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B
|
|
chr1_-_199259133
|
0.554
|
|
KIF21B
|
kinesin family member 21B
|
|
chr6_+_37333484
|
0.551
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr2_+_32435550
|
0.550
|
NM_016252
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr6_-_26305456
|
0.547
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr6_-_26297282
|
0.546
|
NM_003539
|
HIST1H4D
|
histone cluster 1, H4d
|
|
chr9_-_122382079
|
0.545
|
NM_001011649
NM_018249
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2
|
|
chr10_+_86121526
|
0.543
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr7_-_131843377
|
0.542
|
|
PLXNA4
|
plexin A4
|
|
chr22_+_18330049
|
0.542
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr19_+_16039170
|
0.541
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr20_-_33793591
|
0.540
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr17_+_23747422
|
0.539
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr17_-_24645289
|
0.536
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chrX_+_152606945
|
0.536
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr11_-_65962836
|
0.535
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr11_-_5756472
|
0.532
|
NM_001001922
|
OR52N5
|
olfactory receptor, family 52, subfamily N, member 5
|
|
chr19_+_43557264
|
0.528
|
|
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8
|
|
chr1_-_2451543
|
0.528
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr11_-_64302440
|
0.528
|
|
SF1
|
splicing factor 1
|
|
chr6_-_33864883
|
0.527
|
NM_181336
|
LEMD2
|
LEM domain containing 2
|
|
chr6_-_144458326
|
0.527
|
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa
|
|
chr9_+_125158268
|
0.525
|
NM_173689
|
CRB2
|
crumbs homolog 2 (Drosophila)
|
|
chr12_+_48765300
|
0.524
|
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr21_+_42946719
|
0.523
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr12_-_69317441
|
0.523
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr12_+_48765209
|
0.522
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr1_-_114156429
|
0.521
|
NM_018364
|
RSBN1
|
round spermatid basic protein 1
|
|
chr9_-_132803809
|
0.520
|
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr19_-_12905528
|
0.520
|
NM_004461
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr16_-_30841938
|
0.519
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr16_-_30477006
|
0.517
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr6_+_35373572
|
0.516
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr6_-_18095831
|
0.515
|
|
KIF13A
|
kinesin family member 13A
|
|
chr1_-_100370994
|
0.514
|
NM_194292
|
SASS6
|
spindle assembly 6 homolog (C. elegans)
|
|
chr19_+_59077241
|
0.514
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr21_+_33844793
|
0.512
|
|
SON
|
SON DNA binding protein
|
|
chr1_-_198643885
|
0.512
|
|
ZNF281
|
zinc finger protein 281
|
|
chr3_-_12680626
|
0.509
|
NM_002880
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr5_+_140166842
|
0.509
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr6_+_42639929
|
0.508
|
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2
|
|
chr2_+_177785667
|
0.508
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr22_+_21027538
|
0.506
|
|
|
|
|
chr2_+_3620296
|
0.503
|
NM_024027
NM_199235
|
COLEC11
|
collectin sub-family member 11
|
|
chr17_-_46479127
|
0.502
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr6_+_26325124
|
0.502
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr20_+_42309371
|
0.502
|
|
GDAP1L1
|
ganglioside-induced differentiation-associated protein 1-like 1
|
|
chr20_+_36786518
|
0.502
|
NM_080552
|
SLC32A1
|
solute carrier family 32 (GABA vesicular transporter), member 1
|
|
chr17_+_45221002
|
0.501
|
|
MYST2
|
MYST histone acetyltransferase 2
|
|
chr2_+_170298487
|
0.500
|
|
|
|
|
chr6_+_2990616
|
0.498
|
NM_001012983
|
FAM136BP
|
family with sequence similarity 136, member B, pseudogene
|
|
chr17_-_72244231
|
0.495
|
|
|
|