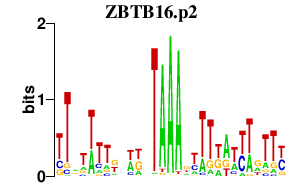
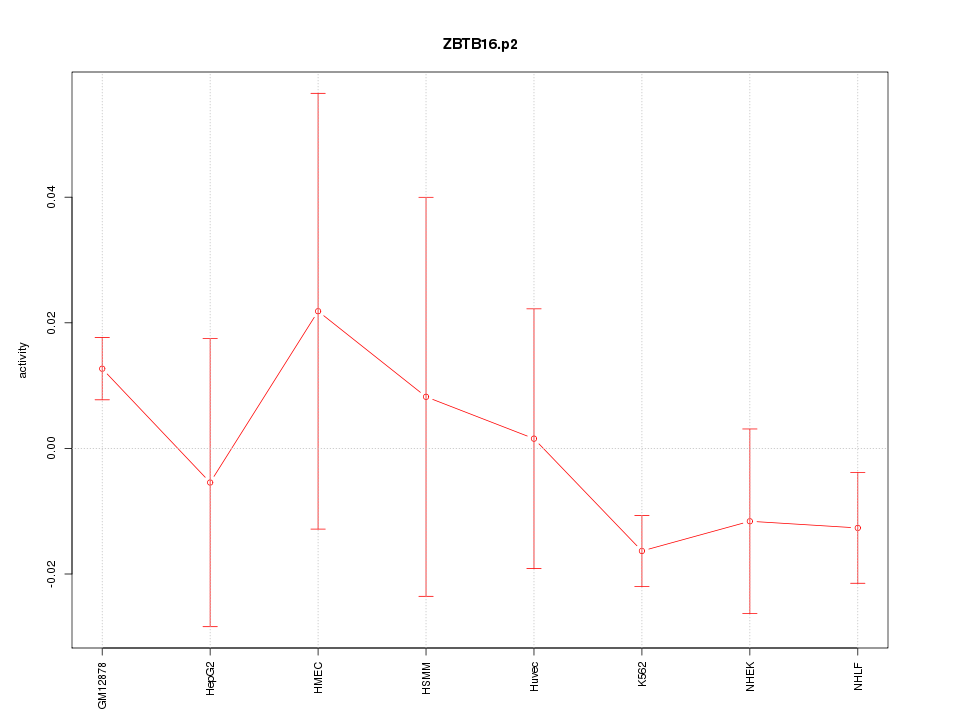
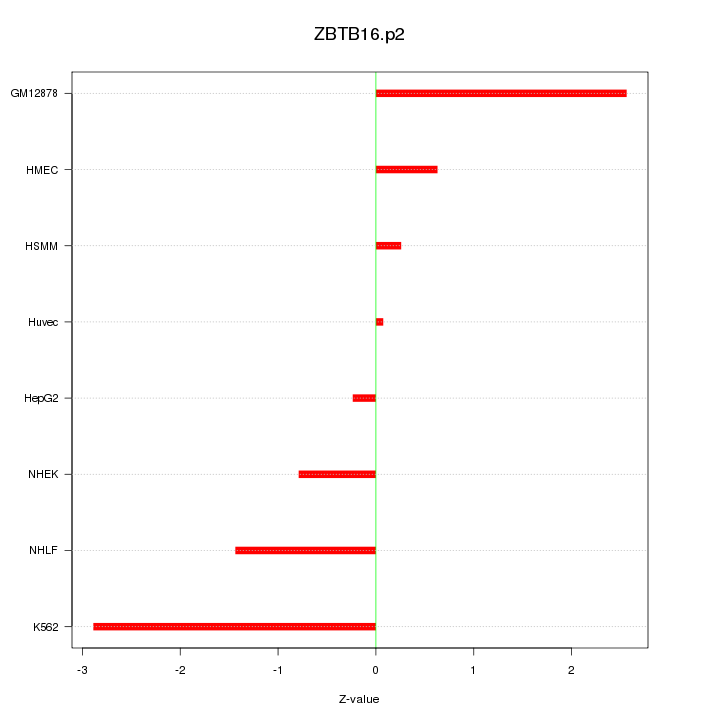
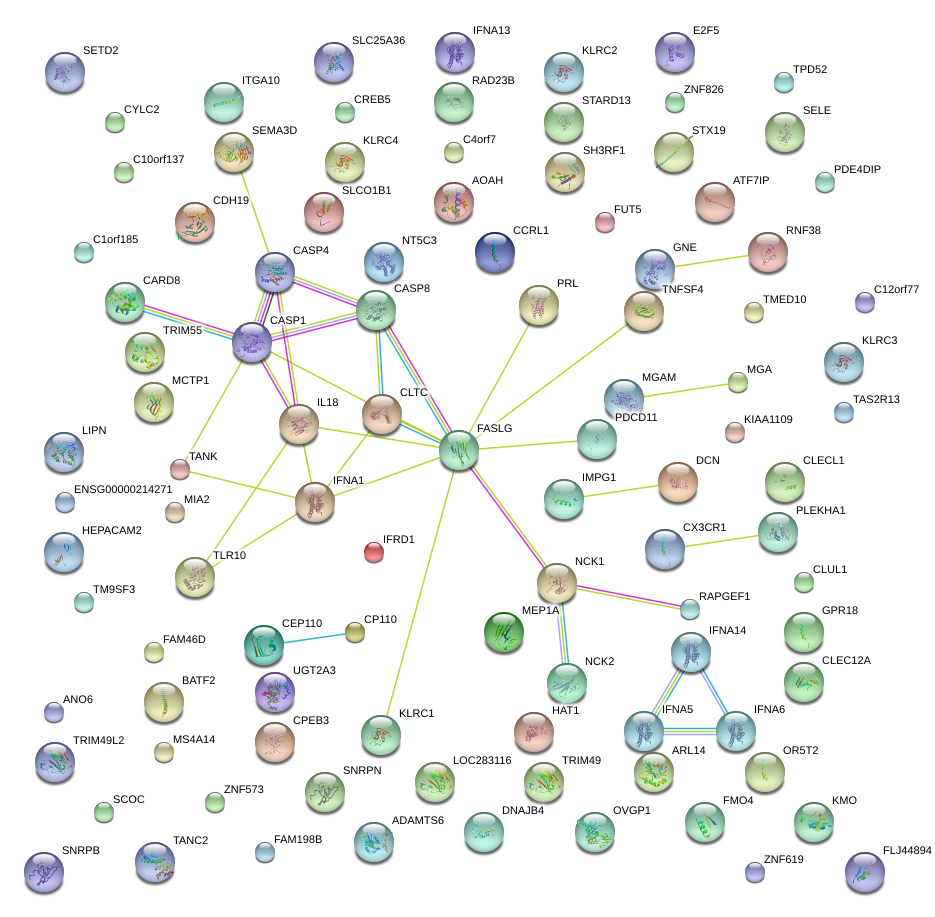
|
chr12_-_10453409
|
5.950
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr11_-_104332631
|
5.438
|
NM_033306
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr12_-_10953427
|
5.069
|
NM_023920
|
TAS2R13
|
taste receptor, type 2, member 13
|
|
chr9_+_104797413
|
5.055
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr19_-_53444733
|
4.339
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr10_-_98315090
|
4.070
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr12_+_14429499
|
4.003
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr4_-_38460983
|
3.433
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr8_-_81155564
|
3.381
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr4_-_69852090
|
3.009
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr15_+_22832198
|
2.949
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr19_-_20399601
|
2.891
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr9_+_21430439
|
2.891
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr11_-_89181390
|
2.788
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr5_-_64813459
|
2.766
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr1_+_144236398
|
2.761
|
|
ITGA10
|
integrin, alpha 10
|
|
chr10_-_93992982
|
2.718
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr9_-_36266930
|
2.643
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr11_+_59920062
|
2.609
|
NM_001079692
NM_032597
|
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14
|
|
chr9_-_21340885
|
2.585
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr4_+_141484046
|
2.560
|
NM_001153446
NM_001153635
NM_001153585
NM_001153690
|
SCOC
|
short coiled-coil protein
|
|
chr1_+_239762179
|
2.547
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr1_+_144236346
|
2.541
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr4_+_71126376
|
2.501
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr14_+_38772870
|
2.493
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr13_-_98708549
|
2.470
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr2_+_161701711
|
2.421
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr1_-_167969802
|
2.374
|
NM_000450
|
SELE
|
selectin E
|
|
chr12_-_9987277
|
2.369
|
|
LOC100506159
|
hypothetical LOC100506159
|
|
chr2_+_201833467
|
2.344
|
NM_033356
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr12_+_21175394
|
2.292
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr3_-_95230143
|
2.255
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr7_-_84589182
|
2.250
|
NM_152754
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr12_-_90096275
|
2.245
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr7_+_111879348
|
2.226
|
NM_001197079
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr1_-_143708467
|
2.213
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_+_63510481
|
2.193
|
|
|
|
|
chr7_-_33047042
|
2.185
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr18_+_606671
|
2.172
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr4_-_159313613
|
2.162
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr19_+_20750949
|
2.087
|
|
|
|
|
chr1_+_51340487
|
2.082
|
NM_001136508
|
C1orf185
|
chromosome 1 open reading frame 185
|
|
chr1_-_111771877
|
2.079
|
NM_002557
|
OVGP1
|
oviductal glycoprotein 1, 120kDa
|
|
chr18_-_62422195
|
1.947
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr17_+_58440629
|
1.945
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr9_-_21229975
|
1.944
|
NM_002172
|
IFNA14
|
interferon, alpha 14
|
|
chr3_-_47073910
|
1.925
|
|
SETD2
|
SET domain containing 2
|
|
chr5_-_94250357
|
1.914
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr9_-_21295311
|
1.903
|
NM_002169
|
IFNA5
|
interferon, alpha 5
|
|
chr8_+_67201684
|
1.900
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr4_+_123311207
|
1.895
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chr11_-_104477367
|
1.883
|
NM_001007232
|
CARD17
|
caspase recruitment domain family, member 17
|
|
chr11_+_89403921
|
1.855
|
NM_001195234
|
TRIM49
TRIM49L2
|
tripartite motif containing 49
tripartite motif containing 49-like 2
|
|
chr9_+_122923864
|
1.839
|
|
CEP110
|
centrosomal protein 110kDa
|
|
chr12_-_9777126
|
1.835
|
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr11_-_111539881
|
1.821
|
NM_001562
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor)
|
|
chr10_+_124141808
|
1.771
|
NM_021622
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chrX_+_79562601
|
1.769
|
NM_152630
|
FAM46D
|
family with sequence similarity 46, member D
|
|
chr7_-_92693716
|
1.754
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr12_-_25041639
|
1.745
|
NM_001101339
|
C12orf77
|
chromosome 12 open reading frame 77
|
|
chr1_+_170894780
|
1.732
|
NM_000639
|
FASLG
|
Fas ligand (TNF superfamily, member 6)
|
|
chr1_-_171441234
|
1.728
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr9_-_133602745
|
1.713
|
NM_005312
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr7_+_28305464
|
1.698
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_-_170314382
|
1.692
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr19_-_5821546
|
1.673
|
NM_002034
|
FUT5
|
fucosyltransferase 5 (alpha (1,3) fucosyltransferase)
|
|
chr8_+_86287161
|
1.670
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr2_+_105799354
|
1.666
|
|
NCK2
|
NCK adaptor protein 2
|
|
chr6_-_76838944
|
1.658
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr14_-_74669864
|
1.655
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr3_+_161877640
|
1.637
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr12_-_10479810
|
1.633
|
NM_002260
|
KLRC2
KLRC3
|
killer cell lectin-like receptor subfamily C, member 2
killer cell lectin-like receptor subfamily C, member 3
|
|
chr6_-_22410993
|
1.620
|
NM_001163558
|
PRL
|
prolactin
|
|
chr13_-_32678142
|
1.603
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_+_55098280
|
1.586
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr12_+_10015274
|
1.584
|
NM_138337
NM_201623
|
CLEC12A
|
C-type lectin domain family 12, member A
|
|
chr11_-_55757236
|
1.583
|
NM_001004746
|
OR5T2
|
olfactory receptor, family 5, subfamily T, member 2
|
|
chr15_+_39839866
|
1.581
|
|
MGA
|
MAX gene associated
|
|
chr9_+_109131814
|
1.578
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr6_+_46869052
|
1.575
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr1_+_169549996
|
1.573
|
NM_002022
|
FMO4
|
flavin containing monooxygenase 4
|
|
chr10_+_90511142
|
1.572
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr7_-_36730525
|
1.556
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr3_+_133798770
|
1.536
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr3_+_138132006
|
1.536
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr3_+_40493605
|
1.529
|
NM_001145082
NM_001145093
NM_001145083
|
ZNF619
|
zinc finger protein 619
|
|
chr21_-_26345294
|
1.518
|
|
|
|
|
chr5_-_74065004
|
1.518
|
|
|
|
|
chr2_+_172530120
|
1.511
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr1_+_78243181
|
1.499
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr3_+_142175329
|
1.458
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr12_+_43972732
|
1.456
|
NM_001142678
|
ANO6
|
anoctamin 6
|
|
chr10_+_127419021
|
1.455
|
|
C10orf137
|
chromosome 10 open reading frame 137
|
|
chr9_-_36365965
|
1.448
|
|
RNF38
|
ring finger protein 38
|
|
chr18_+_30589937
|
1.448
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr11_-_4933518
|
1.441
|
NM_001004748
|
OR51A2
|
olfactory receptor, family 51, subfamily A, member 2
|
|
chr2_+_228064531
|
1.441
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr17_-_36436979
|
1.439
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr17_-_18207416
|
1.426
|
NM_004169
NM_148918
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble)
|
|
chr9_+_674193
|
1.423
|
|
|
|
|
chr2_-_20375497
|
1.412
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr11_-_104411065
|
1.410
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr3_+_180548173
|
1.406
|
NM_033540
|
MFN1
|
mitofusin 1
|
|
chr4_-_17415918
|
1.398
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr3_-_57301749
|
1.393
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr11_-_5212168
|
1.380
|
NM_000519
|
HBD
|
hemoglobin, delta
|
|
chr8_+_23157094
|
1.378
|
NM_152272
|
CHMP7
|
CHMP family, member 7
|
|
chr8_-_37847095
|
1.378
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chrX_+_35847771
|
1.372
|
NM_152632
|
CXorf22
|
chromosome X open reading frame 22
|
|
chr22_-_34564611
|
1.368
|
|
RPL41P5
|
ribosomal protein L41 pseudogene 5
|
|
chr21_-_30774502
|
1.363
|
NM_181607
|
KRTAP19-1
|
keratin associated protein 19-1
|
|
chr19_+_56990222
|
1.357
|
NM_002030
|
FPR3
|
formyl peptide receptor 3
|
|
chr9_-_21197141
|
1.357
|
NM_002171
|
IFNA10
|
interferon, alpha 10
|
|
chr3_+_85858321
|
1.352
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr7_-_18033970
|
1.351
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr4_+_71298187
|
1.348
|
NM_021225
|
PROL1
|
proline rich, lacrimal 1
|
|
chr6_-_109810396
|
1.343
|
NM_001142401
NM_001142402
NM_001142403
NM_001142404
NM_006016
|
CD164
|
CD164 molecule, sialomucin
|
|
chr5_-_75044001
|
1.341
|
NM_152408
|
POC5
|
POC5 centriolar protein homolog (Chlamydomonas)
|
|
chr3_+_99334230
|
1.336
|
NM_001005338
|
OR5H1
|
olfactory receptor, family 5, subfamily H, member 1
|
|
chr12_+_32546243
|
1.334
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr18_-_49944986
|
1.333
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr10_+_96433240
|
1.326
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr11_+_56224436
|
1.319
|
NM_001005213
NM_001013358
|
OR9G1
OR9G9
|
olfactory receptor, family 9, subfamily G, member 1
olfactory receptor, family 9, subfamily G, member 9
|
|
chr2_-_46988449
|
1.314
|
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr10_-_11614279
|
1.310
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr19_+_20138991
|
1.306
|
NM_052852
|
ZNF486
|
zinc finger protein 486
|
|
chr7_-_14847479
|
1.305
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr12_-_15757114
|
1.304
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chrX_-_114158388
|
1.303
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr5_+_156540425
|
1.298
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr12_-_10870134
|
1.295
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chr3_-_158700036
|
1.292
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr11_+_111551417
|
1.289
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr6_-_138581319
|
1.275
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr14_+_88130482
|
1.273
|
NM_207662
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr16_+_87811585
|
1.268
|
NM_182531
|
ZNF778
|
zinc finger protein 778
|
|
chr8_-_120720200
|
1.262
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr6_+_130799954
|
1.259
|
NM_052913
|
TMEM200A
|
transmembrane protein 200A
|
|
chr6_+_154449334
|
1.256
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr14_+_38804226
|
1.256
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr7_-_26202626
|
1.256
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr19_-_53912025
|
1.249
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr4_+_69465107
|
1.248
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr21_+_24722913
|
1.240
|
|
FLJ42200
|
FLJ42200 protein
|
|
chr11_-_1586190
|
1.240
|
NM_001012708
|
KRTAP5-3
|
keratin associated protein 5-3
|
|
chr8_-_59575249
|
1.239
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr1_+_114298021
|
1.229
|
NM_181358
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr12_-_10464308
|
1.227
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr3_+_173954940
|
1.226
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr1_+_245779018
|
1.221
|
NM_145278
|
C1orf150
|
chromosome 1 open reading frame 150
|
|
chr1_+_28637247
|
1.220
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr4_-_177427252
|
1.216
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr21_+_30895276
|
1.213
|
NM_181620
|
KRTAP22-1
|
keratin associated protein 22-1
|
|
chrX_+_37777730
|
1.203
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr4_-_64957595
|
1.200
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr1_+_161308318
|
1.195
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_-_52688768
|
1.186
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr5_-_119768430
|
1.181
|
|
|
|
|
chr4_+_113437947
|
1.163
|
NM_001102406
NM_025144
|
ALPK1
|
alpha-kinase 1
|
|
chr9_+_107464558
|
1.162
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr10_+_5126556
|
1.152
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_-_100003936
|
1.145
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr10_-_5436781
|
1.143
|
NM_001171864
NM_024803
|
TUBAL3
|
tubulin, alpha-like 3
|
|
chr20_-_31055892
|
1.141
|
NM_080675
|
SUN5
|
Sad1 and UNC84 domain containing 5
|
|
chr19_-_21438525
|
1.138
|
|
|
|
|
chr10_-_32707549
|
1.138
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr2_-_232034638
|
1.136
|
|
NCL
|
nucleolin
|
|
chr7_+_139750311
|
1.131
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr8_-_114458557
|
1.127
|
NM_198124
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr5_+_140546839
|
1.122
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr1_+_149275652
|
1.119
|
NM_001159642
NM_138278
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like
|
|
chr3_+_46258875
|
1.110
|
NM_001164680
NM_001837
NM_178328
NM_178329
|
CCR3
|
chemokine (C-C motif) receptor 3
|
|
chr3_+_99288706
|
1.109
|
NM_054106
|
OR5AC2
|
olfactory receptor, family 5, subfamily AC, member 2
|
|
chr1_+_76024466
|
1.103
|
NM_004582
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit
|
|
chr10_+_5126613
|
1.103
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr7_+_37689903
|
1.102
|
|
|
|
|
chr19_-_19793512
|
1.101
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr6_-_132980901
|
1.099
|
NM_014626
|
TAAR2
|
trace amine associated receptor 2
|
|
chr13_+_48582389
|
1.099
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr19_-_14800275
|
1.089
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr19_+_57540493
|
1.089
|
NM_173530
|
ZNF610
|
zinc finger protein 610
|
|
chr4_+_24844749
|
1.089
|
NM_018323
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta
|
|
chr13_+_22653059
|
1.089
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr1_+_42934910
|
1.082
|
|
YBX1
|
Y box binding protein 1
|
|
chr22_-_15682555
|
1.075
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr3_-_49372584
|
1.074
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr20_-_44494961
|
1.070
|
|
ELMO2
|
engulfment and cell motility 2
|
|
chr6_+_64289585
|
1.066
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr12_-_55990512
|
1.066
|
NM_014925
|
R3HDM2
|
R3H domain containing 2
|
|
chr1_+_173111306
|
1.065
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr7_-_107558021
|
1.063
|
NM_007356
|
LAMB4
|
laminin, beta 4
|
|
chr20_-_43601542
|
1.061
|
NM_080827
|
WFDC6
|
WAP four-disulfide core domain 6
|
|
chr10_+_5126589
|
1.058
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr4_-_47709717
|
1.057
|
NM_000087
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr11_+_17329714
|
1.056
|
|
DKFZp686O24166
|
hypothetical protein DKFZp686O24166
|
|
chr3_+_142587973
|
1.054
|
|
|
|
|
chr6_-_52060327
|
1.050
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr7_+_111908143
|
1.047
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr8_+_24297742
|
1.046
|
NM_001145271
NM_001145272
NM_014479
|
ADAMDEC1
|
ADAM-like, decysin 1
|
|
chr2_-_73373956
|
1.037
|
NM_001965
|
EGR4
|
early growth response 4
|