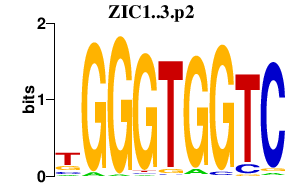
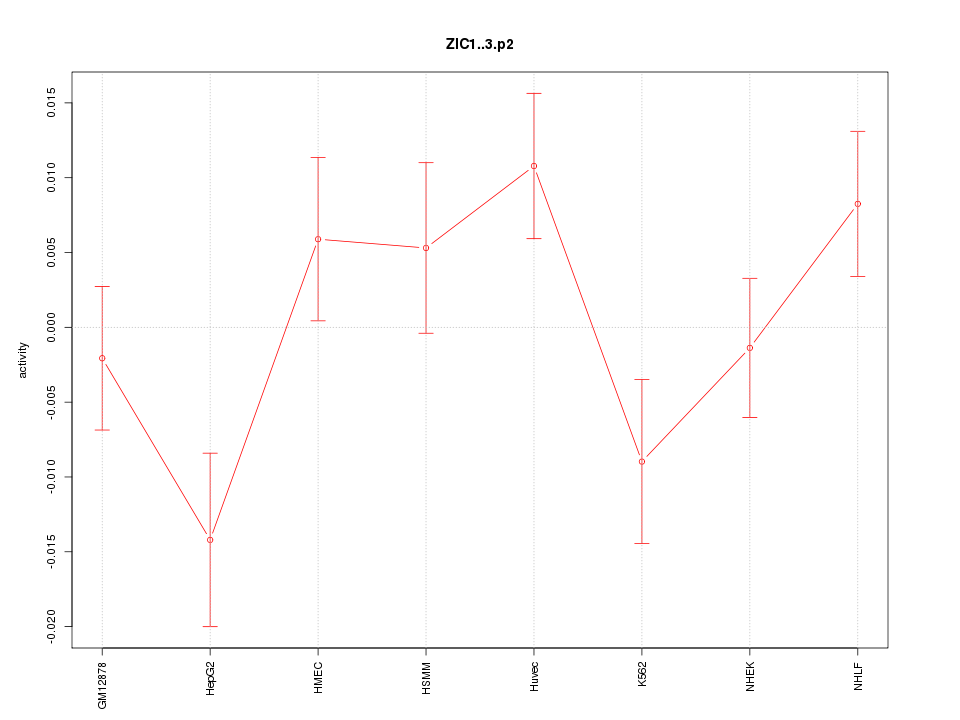
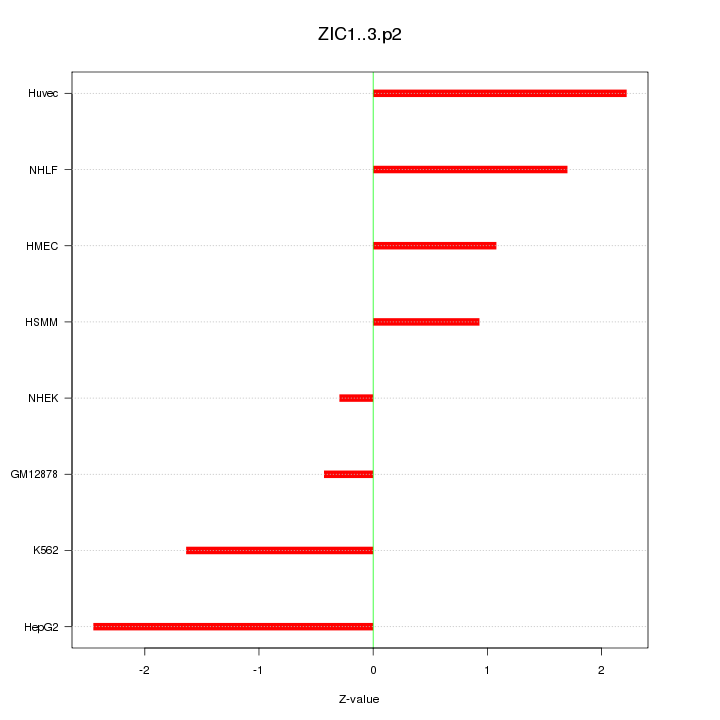
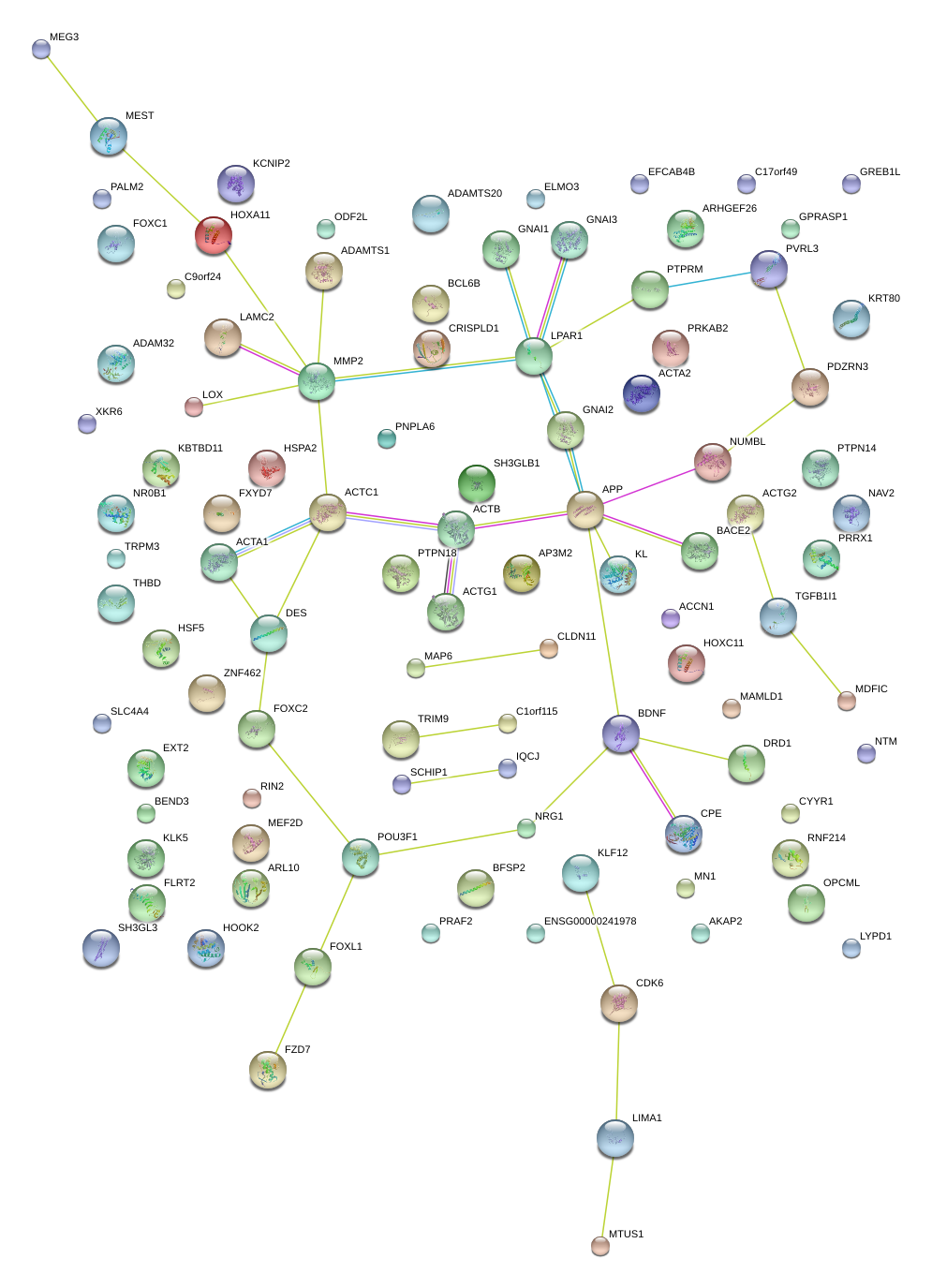
|
chr3_-_73756668
|
2.235
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr3_+_64645819
|
1.418
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr9_+_108665168
|
1.300
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr7_-_106088565
|
1.239
|
NM_175884
|
FLJ36031
|
hypothetical protein FLJ36031
|
|
chr10_-_90702490
|
1.196
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr5_-_174803768
|
1.101
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr21_-_27139563
|
1.095
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr16_-_31390926
|
1.078
|
|
|
|
|
chr7_+_129918408
|
1.072
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr14_+_64077171
|
1.048
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr3_+_155321826
|
1.023
|
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr12_-_3732614
|
0.993
|
NM_001144958
NM_001144959
NM_032680
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr21_+_41461962
|
0.976
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr16_+_65790514
|
0.951
|
NM_024712
|
ELMO3
|
engulfment and cell motility 3
|
|
chr11_+_131285747
|
0.939
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr21_-_27139143
|
0.937
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr2_+_130846263
|
0.928
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr12_-_3732478
|
0.898
|
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr14_+_64076916
|
0.894
|
NM_021979
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr11_+_44074322
|
0.887
|
NM_000401
|
EXT2
|
exostosin 2
|
|
chr17_-_53920699
|
0.827
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr16_+_85158357
|
0.814
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr1_+_168899936
|
0.812
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr1_-_86634511
|
0.796
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chr5_-_121440704
|
0.793
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr22_-_26527469
|
0.784
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr9_+_99656013
|
0.782
|
|
|
|
|
chrX_-_30237403
|
0.782
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr5_+_175725050
|
0.757
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr9_+_111850698
|
0.736
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr1_-_154727014
|
0.729
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr2_+_219991342
|
0.725
|
NM_001927
|
DES
|
desmin
|
|
chr12_-_50871953
|
0.697
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr8_-_11096257
|
0.694
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr19_-_12747421
|
0.672
|
NM_001100176
NM_013312
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr13_+_32488477
|
0.671
|
NM_004795
|
KL
|
klotho
|
|
chr17_-_28644118
|
0.666
|
NM_183377
|
ACCN1
|
amiloride-sensitive cation channel 1, neuronal
|
|
chr16_+_31390567
|
0.662
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr3_+_160964574
|
0.661
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_19691397
|
0.658
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr21_-_26867138
|
0.655
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr19_-_12747263
|
0.651
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr20_-_22978141
|
0.650
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr2_-_133144094
|
0.648
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr17_+_6867090
|
0.620
|
NM_181844
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr9_-_72673753
|
0.617
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr14_+_100365162
|
0.609
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr4_+_166519392
|
0.609
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr1_-_29321599
|
0.609
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr15_+_81906983
|
0.601
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chrX_+_101792949
|
0.599
|
NM_001099410
NM_001099411
NM_014710
NM_001184727
|
GPRASP1
|
G protein-coupled receptor associated sorting protein 1
|
|
chr11_-_75057299
|
0.591
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr8_+_76059262
|
0.579
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr8_+_31616809
|
0.576
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr8_+_42129620
|
0.572
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr3_+_134601479
|
0.565
|
NM_003571
|
BFSP2
|
beaded filament structural protein 2, phakinin
|
|
chr3_+_50272648
|
0.557
|
|
|
|
|
chr2_+_202607554
|
0.557
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr16_+_85169615
|
0.554
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr17_+_6858815
|
0.553
|
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr17_+_6867138
|
0.552
|
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr8_+_1937711
|
0.550
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr13_-_73605914
|
0.549
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr8_+_39084206
|
0.547
|
NM_145004
|
ADAM32
|
ADAM metallopeptidase domain 32
|
|
chr12_-_42231990
|
0.535
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr1_+_181421796
|
0.530
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr3_+_171619346
|
0.528
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr7_+_114350144
|
0.526
|
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr1_+_181422016
|
0.525
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_38285033
|
0.525
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr11_-_27697768
|
0.524
|
NM_001143807
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr7_-_92301147
|
0.521
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr1_+_218930542
|
0.519
|
|
C1orf115
|
chromosome 1 open reading frame 115
|
|
chr11_-_132318741
|
0.519
|
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr1_-_145110669
|
0.518
|
NM_005399
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit
|
|
chr18_+_7557313
|
0.517
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr19_-_56147971
|
0.510
|
NM_001077491
NM_001077492
NM_012427
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr4_+_72271218
|
0.508
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_-_92982927
|
0.502
|
|
|
|
|
chr8_-_17599438
|
0.500
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr16_+_54070302
|
0.498
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_+_181421984
|
0.497
|
|
LAMC2
|
laminin, gamma 2
|
|
chr18_+_17076166
|
0.496
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr8_+_32525269
|
0.485
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr20_+_19903778
|
0.484
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_-_48902692
|
0.482
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr14_+_85066265
|
0.482
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr19_+_40325993
|
0.480
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chrX_+_149282097
|
0.478
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr11_+_116608447
|
0.478
|
NM_207343
NM_001077239
|
RNF214
|
ring finger protein 214
|
|
chr6_+_1556543
|
0.478
|
|
FOXC1
|
forkhead box C1
|
|
chr12_+_52653176
|
0.476
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr12_+_6517600
|
0.475
|
|
|
|
|
chr10_+_119292699
|
0.474
|
|
|
|
|
chr9_-_34387786
|
0.472
|
NM_032596
|
C9orf24
|
chromosome 9 open reading frame 24
|
|
chr6_-_107542328
|
0.471
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr3_+_112273352
|
0.467
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr9_-_112840064
|
0.467
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr7_+_79602023
|
0.467
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chrX_-_48818585
|
0.466
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr19_-_45888259
|
0.465
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr21_+_41461924
|
0.462
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr7_-_27191287
|
0.461
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr1_+_86942874
|
0.460
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr21_-_26465005
|
0.460
|
NM_000484
NM_001136129
NM_001136130
NM_201413
NM_201414
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr10_-_103593529
|
0.459
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr1_+_86942810
|
0.456
|
NM_016009
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr21_-_26464790
|
0.456
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr21_-_26464931
|
0.452
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr1_+_86943074
|
0.451
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr19_+_7505590
|
0.449
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr21_-_26464915
|
0.448
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr14_-_50631407
|
0.445
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr5_-_156935283
|
0.444
|
|
|
|
|
chr1_-_212791188
|
0.443
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr11_+_124115051
|
0.439
|
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr4_-_140696740
|
0.439
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr5_-_138758783
|
0.436
|
NM_001161546
|
LOC389333
|
hypothetical protein LOC389333
|
|
chr13_-_74953873
|
0.432
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr8_-_97242119
|
0.430
|
NM_001001557
|
GDF6
|
growth differentiation factor 6
|
|
chrX_+_90576252
|
0.428
|
NM_080832
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5
|
|
chr11_-_75057062
|
0.426
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr3_+_49566901
|
0.421
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr5_+_140835788
|
0.421
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chrX_-_134013644
|
0.421
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr16_+_51722285
|
0.420
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr3_+_49034494
|
0.419
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr7_+_27190688
|
0.419
|
|
|
|
|
chrX_+_151991474
|
0.418
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr12_+_52733927
|
0.417
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr5_-_168660293
|
0.415
|
NM_003062
|
SLIT3
|
slit homolog 3 (Drosophila)
|
|
chr19_+_60844093
|
0.414
|
NM_207115
|
ZNF580
|
zinc finger protein 580
|
|
chr5_+_140835727
|
0.410
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr11_+_13646822
|
0.410
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr10_+_68355797
|
0.408
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr17_-_15185623
|
0.407
|
NM_031898
|
TEKT3
|
tektin 3
|
|
chr1_+_148788410
|
0.407
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr12_-_21985433
|
0.406
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr7_-_27180398
|
0.404
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr9_-_122516399
|
0.403
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr6_-_128883125
|
0.403
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr1_+_64709063
|
0.403
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr4_+_159311353
|
0.401
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr1_+_86943103
|
0.399
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr2_-_192767844
|
0.399
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr16_+_66244314
|
0.396
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr9_+_111850770
|
0.395
|
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr14_+_100362236
|
0.395
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr6_+_101948102
|
0.395
|
|
|
|
|
chr19_+_7505037
|
0.392
|
NM_006702
NM_001166112
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr10_-_71575953
|
0.392
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr7_+_27191534
|
0.391
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr17_-_7083321
|
0.390
|
NM_024297
|
PHF23
|
PHD finger protein 23
|
|
chr14_+_102662416
|
0.386
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr22_-_49063088
|
0.385
|
|
PLXNB2
|
plexin B2
|
|
chr14_+_100362204
|
0.385
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_+_124114985
|
0.384
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr10_+_8137356
|
0.380
|
|
GATA3
|
GATA binding protein 3
|
|
chr10_-_105352098
|
0.378
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chrX_-_107866262
|
0.377
|
NM_003604
|
IRS4
|
insulin receptor substrate 4
|
|
chr11_+_13646839
|
0.374
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr7_+_44110527
|
0.372
|
|
AEBP1
|
AE binding protein 1
|
|
chr13_+_41520888
|
0.370
|
NM_152910
NM_178009
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr1_+_110555433
|
0.368
|
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr7_+_77005287
|
0.361
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr10_+_80672842
|
0.359
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr13_+_30378311
|
0.359
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chrX_-_37591760
|
0.359
|
NM_006520
|
DYNLT3
|
dynein, light chain, Tctex-type 3
|
|
chr13_-_109236897
|
0.357
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr11_+_13646806
|
0.356
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr2_-_31214788
|
0.353
|
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chrX_+_51653348
|
0.352
|
NM_001005333
NM_006986
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr20_+_61622985
|
0.352
|
|
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor homolog (zebrafish)
|
|
chr15_+_37660568
|
0.352
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr11_+_122806196
|
0.351
|
|
LOC100128242
|
hypothetical LOC100128242
|
|
chr1_+_153560351
|
0.350
|
NM_001105205
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr6_+_17389752
|
0.349
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_-_37454933
|
0.347
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr4_-_120767777
|
0.346
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr8_-_22044463
|
0.346
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr22_-_17891777
|
0.345
|
|
CLDN5
|
claudin 5
|
|
chr6_-_85529884
|
0.344
|
|
TBX18
|
T-box 18
|
|
chr9_+_4480193
|
0.342
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr7_-_107883929
|
0.341
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr10_-_62373828
|
0.341
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr1_+_100777044
|
0.338
|
|
GPR88
|
G protein-coupled receptor 88
|
|
chr5_-_156935317
|
0.335
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr2_+_182030510
|
0.333
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr4_-_159311959
|
0.332
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr13_+_102047286
|
0.331
|
NM_003291
|
TPP2
|
tripeptidyl peptidase II
|
|
chr7_-_5535349
|
0.331
|
|
ACTB
|
actin, beta
|
|
chr19_-_53912025
|
0.330
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr15_-_27901828
|
0.330
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr12_+_6179736
|
0.328
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chrX_+_104953215
|
0.326
|
|
NRK
|
Nik related kinase
|
|
chr12_+_77782648
|
0.325
|
|
SYT1
|
synaptotagmin I
|
|
chr5_-_58607673
|
0.324
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_+_72653420
|
0.321
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr17_-_602125
|
0.320
|
|
GEMIN4
|
gem (nuclear organelle) associated protein 4
|
|
chr2_-_175255717
|
0.320
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|