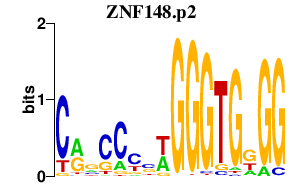
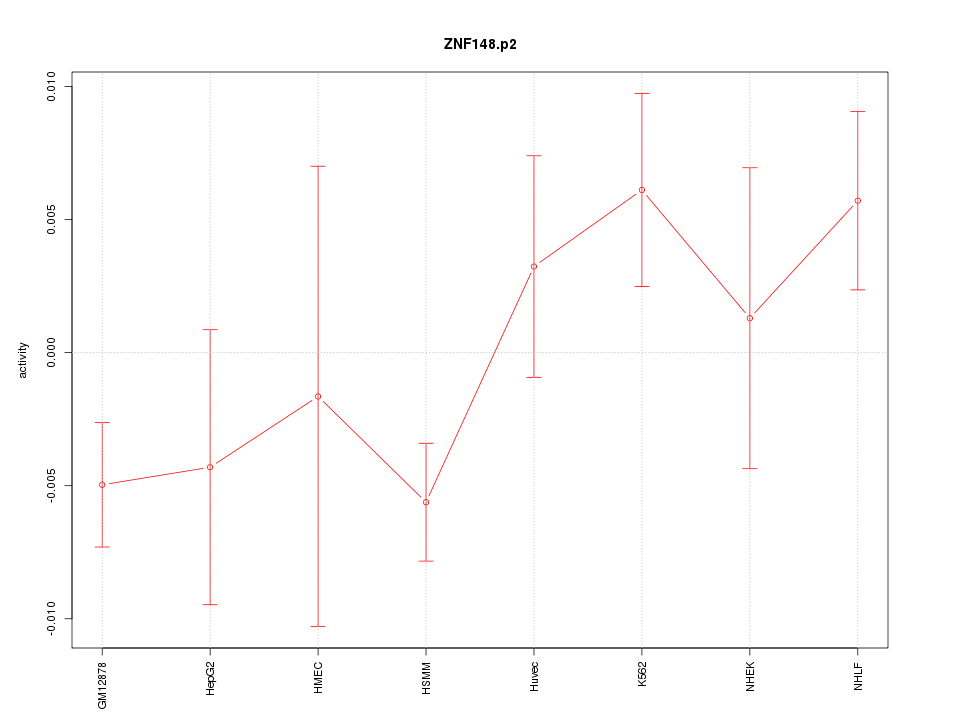
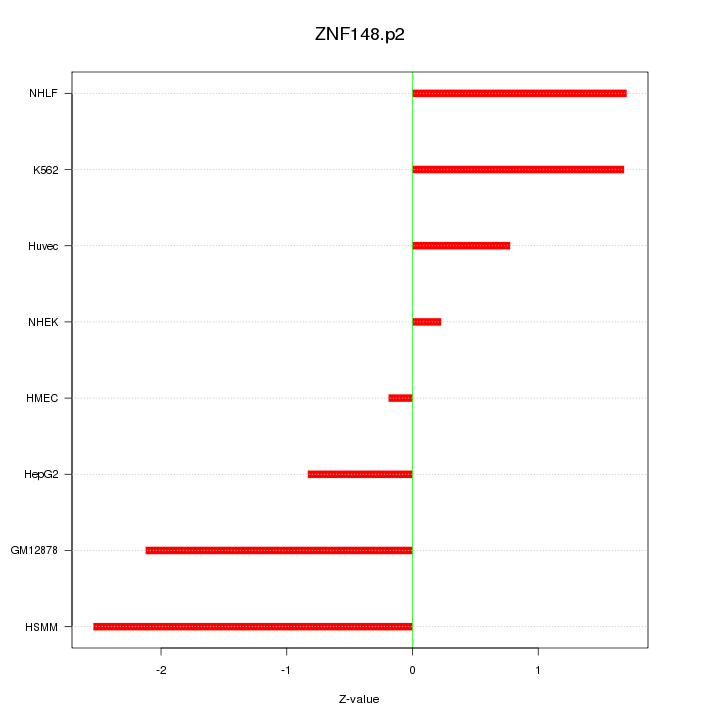
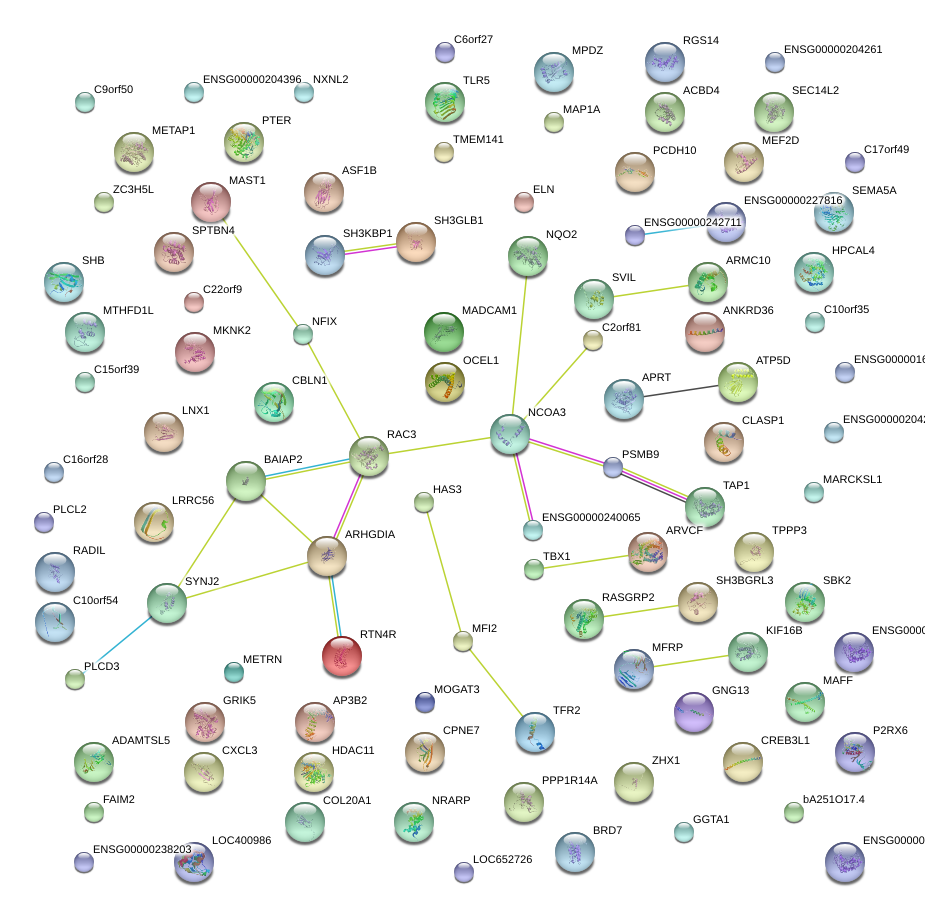
|
chr19_+_12805747
|
1.758
|
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr5_+_176726918
|
1.251
|
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr19_+_45728207
|
1.156
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr19_+_45728270
|
1.119
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr3_+_16901455
|
1.050
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr5_-_9599157
|
0.965
|
NM_003966
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
|
|
chr10_-_135088065
|
0.924
|
NM_001012508
|
SPRN
|
shadow of prion protein homolog (zebrafish)
|
|
chr9_-_131422842
|
0.917
|
NM_199350
|
C9orf50
|
chromosome 9 open reading frame 50
|
|
chr9_+_90339835
|
0.864
|
NM_001161625
NM_145283
|
NXNL2
|
nucleoredoxin-like 2
|
|
chr11_-_118716244
|
0.855
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr6_-_32929570
|
0.851
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr10_-_73203202
|
0.827
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr16_+_705080
|
0.822
|
NM_024042
|
METRN
|
meteorin, glial cell differentiation regulator
|
|
chr9_-_123301856
|
0.819
|
|
GGTA1
|
glycoprotein, alpha-galactosyltransferase 1 pseudogene
|
|
chr19_-_50975121
|
0.810
|
|
|
|
|
chr19_+_13066956
|
0.810
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_-_1997229
|
0.806
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr22_-_43987010
|
0.806
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr6_-_32929721
|
0.792
|
NM_000593
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr16_-_47872973
|
0.788
|
|
CBLN1
|
cerebellin 1 precursor
|
|
chr19_-_1463997
|
0.769
|
|
ADAMTSL5
|
ADAMTS-like 5
|
|
chr4_-_75123193
|
0.762
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr16_+_88169612
|
0.761
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr16_-_87405817
|
0.740
|
NM_000485
NM_001030018
|
APRT
|
adenine phosphoribosyltransferase
|
|
chr10_+_16518947
|
0.725
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr8_+_144888275
|
0.712
|
|
LOC100128338
|
hypothetical LOC100128338
|
|
chr11_+_527521
|
0.709
|
NM_198075
|
LRRC56
|
leucine rich repeat containing 56
|
|
chr6_+_2945025
|
0.706
|
NM_000904
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2
|
|
chr1_+_1062137
|
0.696
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr17_-_40565207
|
0.684
|
NM_133373
|
PLCD3
|
phospholipase C, delta 3
|
|
chr3_+_198214173
|
0.680
|
|
LOC100507057
MFI2
|
hypothetical LOC100507057
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr6_-_32929390
|
0.675
|
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr9_-_38059207
|
0.670
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr9_+_138805637
|
0.669
|
|
TMEM141
|
transmembrane protein 141
|
|
chr7_+_149733309
|
0.662
|
|
|
|
|
chr11_-_64268126
|
0.661
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr16_-_1404635
|
0.659
|
NM_001037125
NM_001193388
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr17_-_77421980
|
0.652
|
NM_001185077
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr1_-_32574185
|
0.651
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr22_-_18635614
|
0.649
|
NM_023004
|
RTN4R
|
reticulon 4 receptor
|
|
chr19_+_447453
|
0.642
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr1_-_39929654
|
0.632
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr22_+_18124225
|
0.628
|
NM_005992
NM_080646
NM_080647
|
TBX1
|
T-box 1
|
|
chr16_-_65984836
|
0.627
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr20_+_61394982
|
0.623
|
NM_020882
|
COL20A1
|
collagen, type XX, alpha 1
|
|
chrX_-_19815526
|
0.617
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_+_134293171
|
0.609
|
|
PCDH10
|
protocadherin 10
|
|
chr16_+_67698943
|
0.603
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr16_-_47872880
|
0.596
|
|
|
|
|
chr15_+_73281286
|
0.593
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr19_-_47261761
|
0.589
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr7_-_4889772
|
0.588
|
NM_018059
|
RADIL
|
Ras association and DIL domains
|
|
chr7_+_102502621
|
0.586
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr15_-_19221463
|
0.585
|
|
|
|
|
chr3_+_13496714
|
0.585
|
NM_024827
NM_001136041
|
HDAC11
|
histone deacetylase 11
|
|
chr6_+_151228383
|
0.581
|
NM_015440
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr15_+_73281249
|
0.571
|
NM_015492
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr16_-_48960186
|
0.568
|
NM_001173984
NM_013263
|
BRD7
|
bromodomain containing 7
|
|
chr22_+_19699441
|
0.564
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr15_+_73281288
|
0.563
|
|
C15orf39
|
chromosome 15 open reading frame 39
|
|
chr19_-_14108267
|
0.559
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr12_-_48583842
|
0.551
|
NM_012306
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr22_+_36928972
|
0.550
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr1_-_154727014
|
0.549
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr9_+_138805581
|
0.546
|
NM_032928
|
TMEM141
|
transmembrane protein 141
|
|
chr10_-_30065760
|
0.544
|
|
SVIL
|
supervillin
|
|
chr15_-_81146188
|
0.543
|
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr6_+_158322916
|
0.540
|
|
SYNJ2
|
synaptojanin 2
|
|
chr19_-_60740246
|
0.540
|
NM_001101401
|
SBK2
|
SH3-binding domain kinase family, member 2
|
|
chr6_+_32929902
|
0.537
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr6_+_158322875
|
0.536
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr7_+_73080362
|
0.535
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr7_+_102502816
|
0.535
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr1_-_1346465
|
0.532
|
NM_001145210
|
LOC441869
|
hypothetical protein LOC441869
|
|
chr2_-_97572685
|
0.531
|
NM_025190
|
ANKRD36B
|
ankyrin repeat domain 36B
|
|
chr1_+_26479030
|
0.530
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr17_+_77582774
|
0.526
|
NM_005052
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3)
|
|
chr19_+_1192742
|
0.525
|
NM_001001975
NM_001687
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr22_-_43986896
|
0.522
|
|
KIAA0930
|
KIAA0930
|
|
chr9_-_13269562
|
0.522
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr22_-_18384251
|
0.522
|
NM_001670
|
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome
|
|
chr6_+_158322904
|
0.521
|
|
SYNJ2
|
synaptojanin 2
|
|
chr16_-_790699
|
0.520
|
NM_016541
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr7_-_100630912
|
0.520
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr8_-_124355670
|
0.517
|
NM_001017926
NM_007222
|
ZHX1
|
zinc fingers and homeoboxes 1
|
|
chr11_+_46256130
|
0.513
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr15_+_41597097
|
0.511
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr19_-_43439011
|
0.506
|
NM_033256
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr19_+_17198012
|
0.503
|
NM_024578
|
OCEL1
|
occludin/ELL domain containing 1
|
|
chr17_+_6858779
|
0.500
|
NM_001142798
NM_001142799
NM_174893
|
C17orf49
|
chromosome 17 open reading frame 49
|
|
chr9_-_139316523
|
0.499
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr19_-_14108400
|
0.499
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr1_+_86943074
|
0.499
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr10_+_71060008
|
0.498
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr19_+_1192766
|
0.496
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr6_-_31853027
|
0.496
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr17_+_40565492
|
0.493
|
NM_001135704
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr7_-_100068612
|
0.491
|
|
TFR2
|
transferrin receptor 2
|
|
chr1_+_86943103
|
0.486
|
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr19_+_1192767
|
0.481
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr17_+_76623540
|
0.480
|
NM_001144888
NM_006340
NM_017450
NM_017451
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr9_-_38059082
|
0.479
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr1_-_221383212
|
0.479
|
NM_003268
|
TLR5
|
toll-like receptor 5
|
|
chr9_+_138991775
|
0.477
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chrX_-_19815370
|
0.476
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr5_-_89805866
|
0.476
|
|
MBLAC2
|
metallo-beta-lactamase domain containing 2
|
|
chr10_-_135232865
|
0.476
|
NM_130784
|
SYCE1
|
synaptonemal complex central element protein 1
|
|
chr6_+_151228577
|
0.474
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr11_-_68537243
|
0.473
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr1_-_42274160
|
0.473
|
|
|
|
|
chr19_+_1192799
|
0.472
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr12_-_112394259
|
0.469
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr1_-_209818675
|
0.464
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr21_-_43369492
|
0.463
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr16_+_2672476
|
0.460
|
NM_018992
|
KCTD5
|
potassium channel tetramerisation domain containing 5
|
|
chr11_+_124240491
|
0.460
|
NM_022370
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila)
|
|
chrX_-_18912365
|
0.459
|
NM_000292
|
PHKA2
|
phosphorylase kinase, alpha 2 (liver)
|
|
chr1_+_26479173
|
0.458
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr7_-_124192916
|
0.457
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr1_+_26479167
|
0.456
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr1_-_39910296
|
0.455
|
NM_032526
|
NT5C1A
|
5'-nucleotidase, cytosolic IA
|
|
chr16_-_47873183
|
0.454
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr11_-_69228286
|
0.453
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr9_+_138991835
|
0.453
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr1_+_26479208
|
0.452
|
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3
|
|
chr4_+_81406765
|
0.452
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr3_-_38666122
|
0.451
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_-_209818576
|
0.451
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr19_+_1192770
|
0.451
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr8_+_145662510
|
0.449
|
NM_145754
|
KIFC2
|
kinesin family member C2
|
|
chr3_-_189354510
|
0.449
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr16_+_66476281
|
0.447
|
NM_198443
|
NRN1L
|
neuritin 1-like
|
|
chr11_+_64538161
|
0.444
|
NM_001667
|
ARL2-SNX15
ARL2
|
ARL2-SNX15 read-through transcript
ADP-ribosylation factor-like 2
|
|
chr10_+_102810904
|
0.444
|
NM_030929
|
KAZALD1
|
Kazal-type serine peptidase inhibitor domain 1
|
|
chr22_+_28609064
|
0.442
|
NM_021090
NM_153050
NM_153051
|
MTMR3
|
myotubularin related protein 3
|
|
chr19_+_1397268
|
0.440
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr3_-_124650081
|
0.440
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr1_-_3653733
|
0.436
|
|
KIAA0495
|
KIAA0495
|
|
chr7_+_102502529
|
0.432
|
NM_001161009
NM_001161010
NM_001161011
NM_001161012
NM_001161013
NM_031905
|
ARMC10
|
armadillo repeat containing 10
|
|
chr5_+_89806436
|
0.429
|
NM_006467
|
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD)
|
|
chr19_+_40337480
|
0.428
|
NM_144779
NM_001164605
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr3_+_185538998
|
0.428
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr3_+_113287864
|
0.427
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr19_+_7505723
|
0.422
|
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr7_+_73080387
|
0.420
|
|
ELN
|
elastin
|
|
chr7_-_100331417
|
0.417
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr7_-_124192265
|
0.416
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr20_+_41569727
|
0.415
|
NM_032107
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila)
|
|
chr22_+_21065134
|
0.413
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr2_+_130846263
|
0.413
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr13_+_23632788
|
0.412
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr7_-_102502250
|
0.409
|
NM_145032
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr21_-_43368963
|
0.408
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr11_-_76058542
|
0.408
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr3_+_160964574
|
0.408
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr19_+_1192811
|
0.407
|
|
ATP5D
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
|
|
chr7_+_102502578
|
0.407
|
|
ARMC10
|
armadillo repeat containing 10
|
|
chr11_+_915899
|
0.405
|
|
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit
|
|
chr21_+_42512271
|
0.405
|
NM_004915
NM_016818
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chrX_-_48212683
|
0.405
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr12_+_47658502
|
0.405
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr3_+_49211936
|
0.402
|
NM_001135197
|
CCDC36
|
coiled-coil domain containing 36
|
|
chr21_-_46473143
|
0.401
|
NM_001001438
NM_001145436
NM_001145437
NM_002340
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr4_-_75183700
|
0.398
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr17_+_1892026
|
0.396
|
NM_080822
|
OVCA2
|
ovarian tumor suppressor candidate 2
|
|
chr8_-_144731498
|
0.395
|
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1
|
|
chr19_-_56215084
|
0.395
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr19_-_53706644
|
0.394
|
|
|
|
|
chr10_-_30388439
|
0.394
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr14_+_104402626
|
0.394
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr5_-_149473127
|
0.394
|
NM_005211
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr22_+_21065202
|
0.394
|
|
|
|
|
chr17_+_76623553
|
0.393
|
|
BAIAP2
|
BAI1-associated protein 2
|
|
chr19_-_51109875
|
0.393
|
NM_001029861
|
NANOS2
|
nanos homolog 2 (Drosophila)
|
|
chr7_+_98314020
|
0.392
|
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr7_+_151353775
|
0.392
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr7_+_90731614
|
0.392
|
|
FZD1
|
frizzled homolog 1 (Drosophila)
|
|
chr5_+_140286485
|
0.389
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr1_-_2129031
|
0.389
|
NM_001146310
|
C1orf86
|
chromosome 1 open reading frame 86
|
|
chr12_-_122584381
|
0.389
|
|
RILPL1
|
Rab interacting lysosomal protein-like 1
|
|
chr7_+_73080404
|
0.388
|
|
ELN
|
elastin
|
|
chr11_+_47386707
|
0.388
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr10_-_43224312
|
0.388
|
NM_001098205
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr19_+_7505590
|
0.388
|
NM_001166111
NM_001166113
|
PNPLA6
|
patatin-like phospholipase domain containing 6
|
|
chr22_+_21042091
|
0.388
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr1_-_1041598
|
0.386
|
NM_017891
|
C1orf159
|
chromosome 1 open reading frame 159
|
|
chr6_-_30762385
|
0.386
|
|
KIAA1949
|
KIAA1949
|
|
chr1_-_6468108
|
0.385
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_-_3517918
|
0.385
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr1_-_38170004
|
0.385
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr7_-_2561638
|
0.384
|
|
BAAT1
|
BRCA1-associated ATM activator 1
|
|
chr19_+_55614006
|
0.384
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr8_-_145640579
|
0.384
|
|
TONSL
|
tonsoku-like, DNA repair protein
|
|
chr6_+_151228757
|
0.382
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr19_+_810673
|
0.381
|
|
CFD
|
complement factor D (adipsin)
|
|
chr1_-_43197123
|
0.381
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr7_+_73080422
|
0.380
|
|
ELN
|
elastin
|
|
chr16_-_46976649
|
0.380
|
NM_003031
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr7_-_102502407
|
0.379
|
NM_001111038
|
FBXL13
|
F-box and leucine-rich repeat protein 13
|
|
chr11_+_46358909
|
0.379
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|