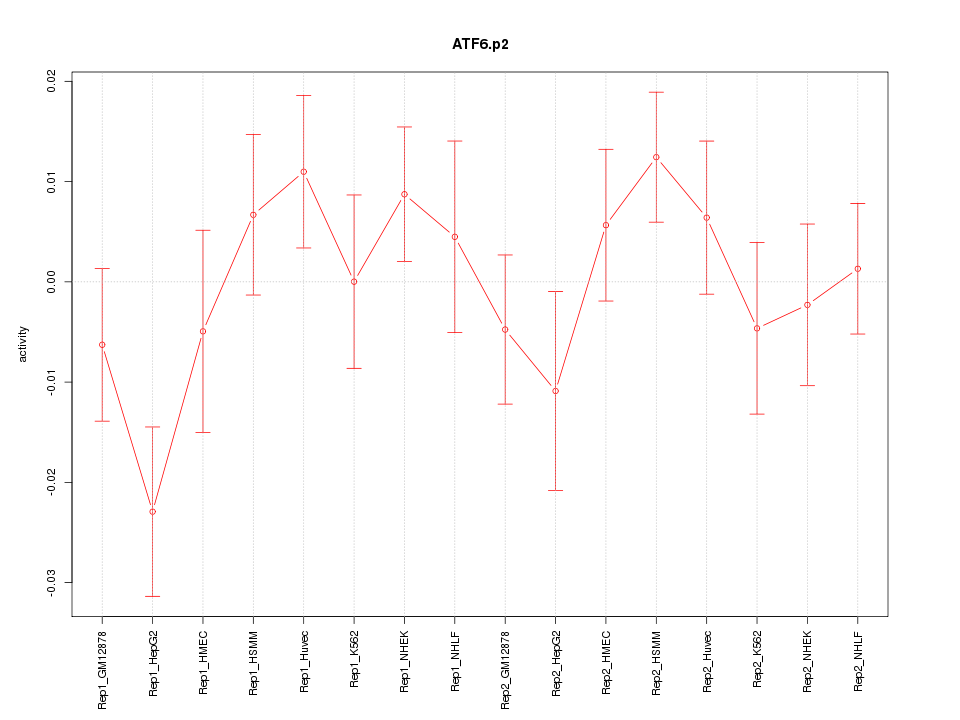
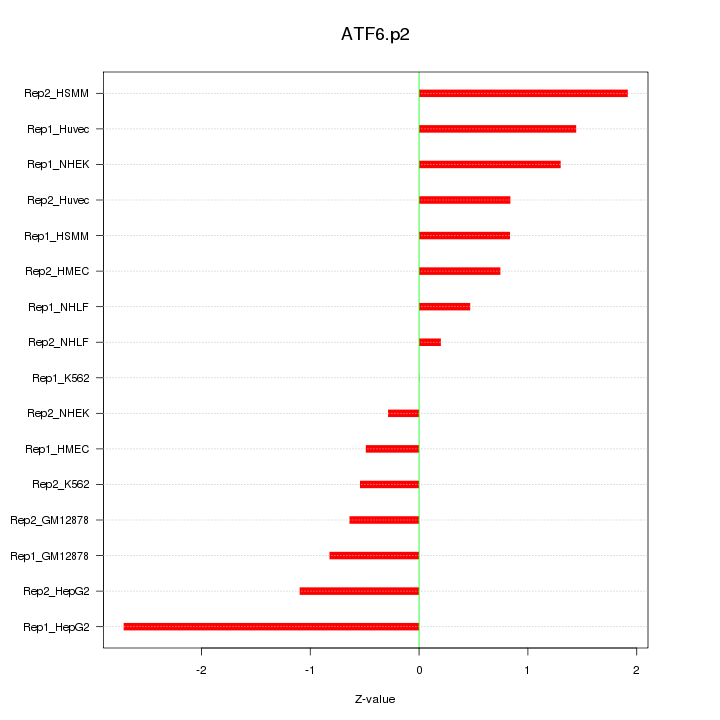
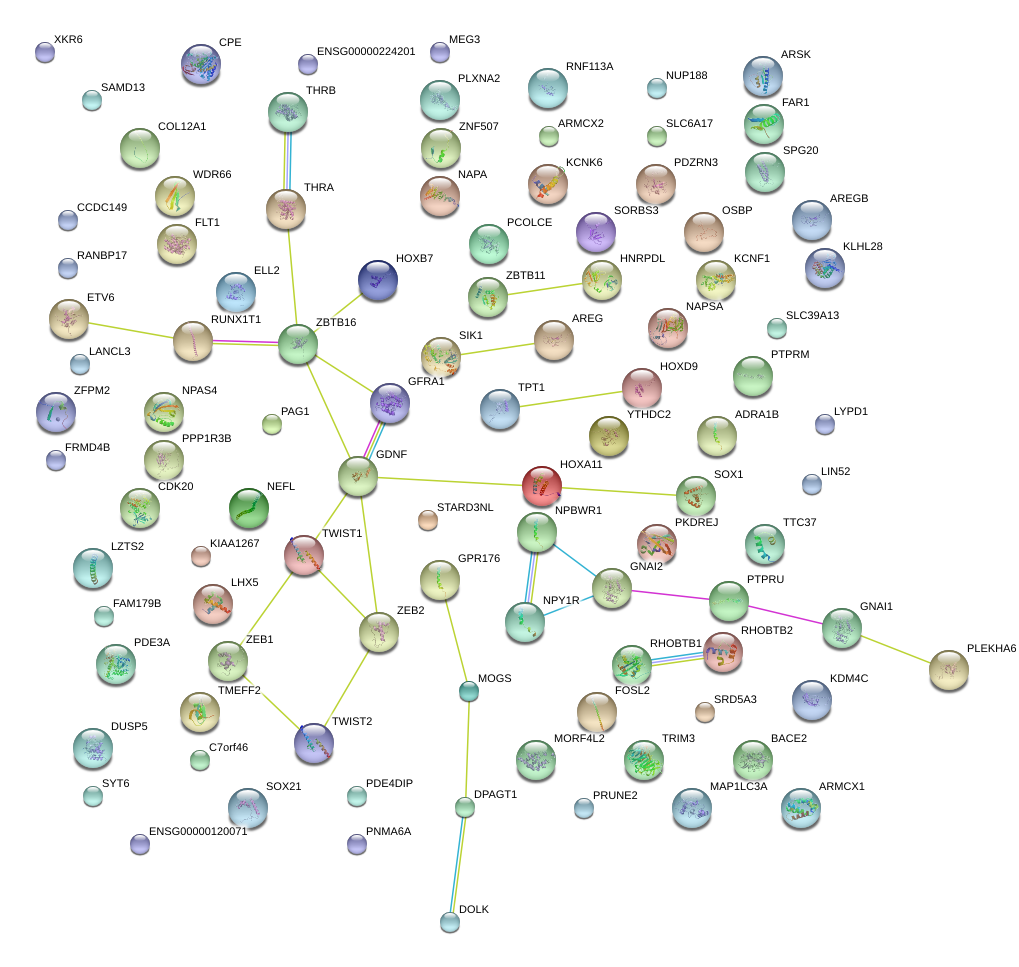
|
chr3_-_73756668
|
1.212
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr1_-_202595666
|
1.093
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr5_-_94916416
|
0.896
|
NM_014639
|
TTC37
|
tetratricopeptide repeat domain 37
|
|
chr5_+_94916533
|
0.853
|
NM_198150
|
ARSK
|
arylsulfatase family, member K
|
|
chr10_-_118022965
|
0.838
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr19_-_52709975
|
0.836
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr19_-_52710089
|
0.809
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr7_+_27191534
|
0.777
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr2_-_192767494
|
0.769
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_-_55532355
|
0.764
|
NM_201286
|
USP51
|
ubiquitin specific peptidase 51
|
|
chr1_+_29435534
|
0.740
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr2_-_133144094
|
0.716
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr19_-_52710129
|
0.713
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr18_+_7557313
|
0.695
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr11_+_113435506
|
0.688
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr4_+_166519392
|
0.683
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr8_+_106400322
|
0.683
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr4_+_75529716
|
0.676
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr7_-_19123765
|
0.676
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr21_-_43671355
|
0.673
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr7_-_19123787
|
0.666
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chrX_-_100801472
|
0.659
|
NM_014782
NM_177949
|
ARMCX2
|
armadillo repeat containing, X-linked 2
|
|
chr11_+_113435640
|
0.651
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_9046460
|
0.650
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr14_+_44501083
|
0.649
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr6_-_75972473
|
0.645
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr22_-_45037882
|
0.630
|
NM_006071
|
PKDREJ
|
polycystic kidney disease (polycystin) and REJ homolog (sperm receptor for egg jelly homolog, sea urchin)
|
|
chr9_-_89779413
|
0.624
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr7_+_23686268
|
0.602
|
NM_001127364
NM_001127365
NM_199136
|
C7orf46
|
chromosome 7 open reading frame 46
|
|
chr4_+_75699652
|
0.601
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr5_-_37875538
|
0.598
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr2_-_192767844
|
0.597
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr9_+_130749812
|
0.595
|
|
NUP188
|
nucleoporin 188kDa
|
|
chr5_+_159276317
|
0.591
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr17_-_41625921
|
0.584
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr1_+_113300507
|
0.582
|
|
LOC100506392
|
hypothetical LOC100506392
|
|
chr10_+_102749597
|
0.575
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr3_-_102878301
|
0.562
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr10_+_112247614
|
0.549
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr13_-_35818396
|
0.543
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr2_+_10969513
|
0.536
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr14_+_73621408
|
0.528
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr8_-_82186832
|
0.516
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr13_-_35818603
|
0.516
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr12_+_20413445
|
0.514
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr9_+_130749791
|
0.512
|
NM_015354
|
NUP188
|
nucleoporin 188kDa
|
|
chr13_-_35818491
|
0.500
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr4_-_164473107
|
0.497
|
NM_000909
|
NPY1R
|
neuropeptide Y receptor Y1
|
|
chr1_-_114497994
|
0.497
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr1_+_110494654
|
0.496
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr2_-_74545917
|
0.494
|
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr3_-_69517913
|
0.493
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr19_+_54996613
|
0.489
|
|
|
|
|
chr13_-_27967215
|
0.488
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr3_-_131313010
|
0.485
|
|
|
|
|
chr3_-_131312963
|
0.485
|
|
|
|
|
chr8_+_22479115
|
0.481
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr5_+_112877251
|
0.481
|
NM_022828
|
YTHDC2
|
YTH domain containing 2
|
|
chr12_+_120840829
|
0.479
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr19_+_43502591
|
0.476
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr9_-_130749604
|
0.472
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr1_-_206484258
|
0.469
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr5_+_170221584
|
0.469
|
NM_022897
|
RANBP17
|
RAN binding protein 17
|
|
chr19_+_43502301
|
0.459
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr14_-_44500871
|
0.457
|
NM_017658
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr13_-_35818777
|
0.453
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr11_-_118477994
|
0.452
|
NM_001382
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr21_+_41461597
|
0.442
|
NM_012105
NM_138991
NM_138992
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr15_-_38000379
|
0.442
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chrX_+_100692071
|
0.438
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr3_+_50259329
|
0.432
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr2_-_74546009
|
0.432
|
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr7_-_27191287
|
0.431
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr8_-_93184629
|
0.427
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_+_11694172
|
0.426
|
|
ETV6
|
ets variant 6
|
|
chr19_+_43502360
|
0.426
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr16_-_31390926
|
0.425
|
|
|
|
|
chrX_-_102828238
|
0.417
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr4_-_187884721
|
0.412
|
|
|
|
|
chr9_-_89779180
|
0.411
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr10_-_62373828
|
0.411
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr1_+_84539876
|
0.410
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr14_-_44500840
|
0.408
|
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr12_-_112394259
|
0.405
|
NM_022363
|
LHX5
|
LIM homeobox 5
|
|
chr20_+_32610161
|
0.402
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr5_+_170221441
|
0.401
|
|
RANBP17
|
RAN binding protein 17
|
|
chr11_+_65945050
|
0.398
|
NM_178864
|
NPAS4
|
neuronal PAS domain protein 4
|
|
chr2_-_74546042
|
0.398
|
NM_001146158
NM_006302
|
MOGS
|
mannosyl-oligosaccharide glucosidase
|
|
chr1_-_143750988
|
0.396
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chrX_+_151991474
|
0.394
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr6_-_75972286
|
0.392
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr13_-_44813277
|
0.391
|
NM_003295
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr2_-_144994038
|
0.388
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr9_-_78710698
|
0.388
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr3_+_50272648
|
0.386
|
|
|
|
|
chr17_-_44043278
|
0.386
|
|
HOXB7
|
homeobox B7
|
|
chr7_+_38184375
|
0.385
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr13_+_44813479
|
0.377
|
|
LOC100190939
|
hypothetical LOC100190939
|
|
chr7_+_38184393
|
0.377
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr8_-_24869945
|
0.376
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr19_+_43502380
|
0.376
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr19_+_37528340
|
0.375
|
NM_001136156
NM_014910
|
ZNF507
|
zinc finger protein 507
|
|
chr4_+_55907138
|
0.373
|
NM_024592
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr5_-_95323353
|
0.372
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr11_-_67329246
|
0.371
|
|
LOC645332
|
family with sequence similarity 86, member A pseudogene
|
|
chr4_-_24523605
|
0.371
|
NM_001130726
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr11_+_47386707
|
0.369
|
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr11_-_6451561
|
0.365
|
NM_006458
NM_033278
|
TRIM3
|
tripartite motif containing 3
|
|
chr7_+_100041143
|
0.365
|
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr9_+_6747919
|
0.362
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr8_+_54015020
|
0.357
|
NM_005285
|
NPBWR1
|
neuropeptides B/W receptor 1
|
|
chr3_-_69518119
|
0.356
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr13_-_94162249
|
0.355
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr7_+_38184363
|
0.355
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr11_+_47386621
|
0.353
|
NM_001128225
NM_152264
|
SLC39A13
|
solute carrier family 39 (zinc transporter), member 13
|
|
chr4_-_83569853
|
0.352
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr17_-_44042924
|
0.351
|
|
HOXB7
|
homeobox B7
|
|
chr3_-_24511456
|
0.351
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr11_+_13646806
|
0.351
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr12_-_123568781
|
0.350
|
|
|
|
|
chr2_+_176695658
|
0.350
|
NM_014213
|
HOXD9
|
homeobox D9
|
|
chr14_+_100362204
|
0.349
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_-_59140192
|
0.346
|
NM_002556
|
OSBP
|
oxysterol binding protein
|
|
chr8_-_11096257
|
0.343
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chrX_+_37315934
|
0.343
|
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr2_+_28469199
|
0.342
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr20_-_4177519
|
0.342
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr6_-_128883125
|
0.341
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr8_+_145718702
|
0.341
|
|
|
|
|
chr11_-_75599433
|
0.339
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr1_-_113300138
|
0.337
|
NM_001166496
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr1_-_113300351
|
0.337
|
NM_003051
|
SLC16A1
|
solute carrier family 16, member 1 (monocarboxylic acid transporter 1)
|
|
chr15_-_42274529
|
0.336
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr13_-_44813308
|
0.335
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr13_-_44813203
|
0.335
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr3_-_188946168
|
0.334
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr7_+_100302885
|
0.333
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr12_+_70434927
|
0.333
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr11_+_66367458
|
0.330
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr1_-_233879617
|
0.328
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr10_+_70553983
|
0.328
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr16_-_4837266
|
0.328
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr11_+_13646822
|
0.326
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr4_+_55907213
|
0.326
|
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr2_+_28469172
|
0.325
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_12692938
|
0.323
|
NM_001037164
NM_212460
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr15_-_70863110
|
0.323
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr4_-_186693574
|
0.321
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr15_+_76952356
|
0.321
|
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr11_+_13646727
|
0.320
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr1_+_32777358
|
0.317
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr11_+_69165065
|
0.317
|
|
CCND1
|
cyclin D1
|
|
chrX_+_37315740
|
0.316
|
NM_001170331
NM_198511
|
LANCL3
|
LanC lantibiotic synthetase component C-like 3 (bacterial)
|
|
chr6_-_91063181
|
0.315
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr15_-_70863115
|
0.314
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr9_+_35528628
|
0.312
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_-_119976631
|
0.311
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr1_+_13899272
|
0.310
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr8_-_29263714
|
0.308
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr11_+_69165119
|
0.308
|
|
CCND1
|
cyclin D1
|
|
chr11_+_69165018
|
0.308
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr15_-_70863124
|
0.307
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr4_-_83569572
|
0.307
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr4_-_83569476
|
0.305
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr9_+_136673464
|
0.305
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr10_+_24023680
|
0.302
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr15_-_70863078
|
0.302
|
|
ADPGK
|
ADP-dependent glucokinase
|
|
chr11_-_65138217
|
0.300
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr1_-_35269836
|
0.300
|
|
ZMYM6
|
zinc finger, MYM-type 6
|
|
chr15_-_66511488
|
0.299
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr19_+_54149843
|
0.299
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr5_-_158459006
|
0.297
|
|
EBF1
|
early B-cell factor 1
|
|
chr9_+_124742932
|
0.295
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr10_+_70553993
|
0.295
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr9_-_126573349
|
0.294
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr13_-_100125028
|
0.294
|
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr13_-_100125103
|
0.293
|
NM_001079669
NM_032813
|
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4
|
|
chr11_+_18676913
|
0.290
|
NM_153347
|
TMEM86A
|
transmembrane protein 86A
|
|
chr4_-_111763356
|
0.288
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr3_-_24511180
|
0.288
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr3_+_124268545
|
0.288
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr14_+_100362236
|
0.285
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_-_122951200
|
0.284
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr19_-_60587414
|
0.283
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr19_-_43438818
|
0.282
|
|
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A
|
|
chr19_-_51608665
|
0.280
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr1_-_184916044
|
0.280
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr19_-_45888259
|
0.280
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr6_+_147871477
|
0.279
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr11_+_18676857
|
0.279
|
|
TMEM86A
|
transmembrane protein 86A
|
|
chr10_+_23768203
|
0.278
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr1_-_1274618
|
0.278
|
|
DVL1
|
dishevelled, dsh homolog 1 (Drosophila)
|
|
chr3_+_150330060
|
0.277
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr8_+_98725505
|
0.277
|
NM_178812
|
MTDH
|
metadherin
|
|
chr10_-_7491204
|
0.276
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr15_+_41871809
|
0.274
|
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 read-through transcript
small EDRK-rich factor 2
|
|
chr19_+_54558798
|
0.274
|
NM_001197302
NM_001197301
NM_014419
|
DKKL1
|
dickkopf-like 1
|
|
chr5_+_102229612
|
0.274
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr13_-_101852028
|
0.274
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr9_+_6747636
|
0.271
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|